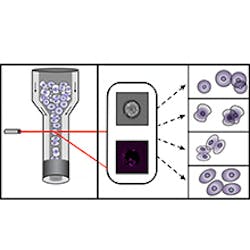
Imaging cytometry technique optimizes cell cycle analysis
Using imaging cytometry data, an international team of scientists at Helmholtz Zentrum München (Neuherberg, Germany) and colleagues has improved the identification of cell cycle phases. The work could eliminate the use of stains by applying algorithms from machine learning. With the help of imaging software, they extracted hundreds of features from brightfield and darkfield images—using this data, they could generate algorithms that can sort the cells digitally.
Related: Instrumentation advances add flexibility and quantitation to flow cytometry
So far, fluorescent stains have been used to assign cells to their cell cycle phase, but these chemicals damage cells and may distort results. The Helmholtz Zentrum München scientists, in collaboration with the Broad Institute of the Massachusetts Institute of Technology (MIT) and Harvard University (both in Cambridge, MA), Swansea University (Wales), Newcastle University (England), and The Francis Crick Institute (England), discovered that brightfield and darkfield images are generally neglected, according to Thomas Blasi, a PhD student at the Broad Institute and first author of the paper describing the work.
By using the information in the data from these images in machine learning, Blasi explains, their approach makes it possible to not only classify cells, but also to digitally sort them with a high level of specificity.
Based on the team's findings, the Broad Institute and Helmholtz Zentrum München have filed a provisional patent application.
Full details of the work appear in the journal Nature Communications; for more information, please visit http://dx.doi.org/10.1038/ncomms10256.
Follow us on Twitter, 'like' us on Facebook, connect with us on Google+, and join our group on LinkedIn