Prior drug development findings for SARS could be effective for Coronavirus
A team of researchers at the University of Chicago (Chicago, IL), the U.S. Department of Energy’s (DOE) Argonne National Laboratory (Lemont, IL), Northwestern University's Feinberg School of Medicine (also in Chicago), and the University of California, Riverside School of Medicine (UCR; Riverside, CA) used a light-based technique to identify a new protein structure of SARS-CoV-2, the virus that causes Coronavirus Disease 2019 (COVID-19). The discovery shows potential as a drug target for the virus.
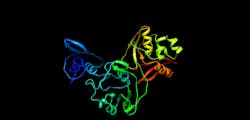
The protein Nsp15 from Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is 89% identical to the protein from the earlier outbreak of SARS-CoV. Studies published in 2010 on SARS-CoV revealed that inhibition of Nsp15 can slow viral replication. This suggests drugs designed to target Nsp15 could be developed as effective drugs against COVID-19.
This new structure was solved by the group of Andrzej Joachimiak, Distinguished Fellow of Argonne, the University of Chicago professor and director of the Structural Biology Center at Argonne’s Advanced Photon Source (APS), a DOE Office of Science user facility in conjunction with the Center for Structural Genomics of Infectious Diseases, of which he is a co-director.
Karla Satchell, principal investigator for the Center for Structural Genomics of Infectious Diseases and professor of microbiology-immunology at Northwestern University, is leading the scientists in investigating the structure of the SARS CoV-2 virus to understand how to stop it from replicating.
The initial genome analysis and design of constructs for protein synthesis were performed by the bioinformatic group of Adam Godzik, a professor of biomedical sciences in the UCR. Mapping a 3D protein structure of the virus, also called solving the structure, allows scientists to figure out how to interfere in the pathogen’s replication in human cells.
“The NSP15 protein has been investigated in SARS as a novel target for new drug development, but that never went very far because the SARS epidemic went away, and all new drug development ended. Some inhibitors were identified, but never developed into drugs. The inhibitors that were developed for SARS now could be tested against this protein,” Satchell says.
Rapid upsurge and proliferation of SARS-CoV-2 raised questions about how this virus could became so much more transmissible as compared to the SARS and MERS coronaviruses. The scientists are mapping the proteins to address this issue.
“While the SARS-CoV-2 is very similar to the SARS virus that caused epidemics in 2003, new structures shed light on the small, but potentially important differences between the two viruses that contribute to the different patterns in the spread and severity of the diseases they cause,” Godzik said.
Satchell, Joachimiak, and Godzik—along with the entire center team—will map the structure of some of the 28 proteins in the virus in order to see where drugs can throw a chemical monkey wrench into its machinery. The proteins are folded globular structures with precisely defined function and their “active sites” can be targeted with chemical compounds.
The first step is to clone and express the genes of the virus proteins and grow them as protein crystals in miniature ice cube-like trays. The consortium includes nine labs across eight institutions that will participate in this effort.
Viewing these proteins down to the arrangement of their atoms requires an intense x-ray beam. Thus, once the crystals are grown, the center scientists image them using the APS’ extremely bright light source in a process called x-ray crystallography. Data for structure determination were collected at Structural Biology Beamlines funded by DOE Office of Biological and Environmental Research.
Satchell and her team are well prepared for the challenges that come with developing drugs to fight the virus. The Center for Structural Genomics of Infectious Diseases, established in 2007, has mapped more than a thousand parts of lethal bacteria and viruses in three dimensions, exposing an intimate chemical portrait of diseases. This view offers scientists a window into the bacteria or virus’ vulnerabilities enables them to create drugs to disable it or vaccines to prevent it.
The structure of Nsp15 was released to the scientific community on March 4, 2020, on the RSCB Protein Data Bank.
Full details of the work appear in bioRxiv.