Laser technique uses diamond defects to reveal protein structures in great detail
Proteins have highly complex structures that could be key to developing drugs or to understanding basic biological processes. But figuring out the arrangement of atoms in these complicated, folded molecules usually requires getting them to form crystals large enough to be observed in detail—and for many proteins, that is either impossible or dauntingly difficult.
Related: Optical method can detect proteins by their scattered light
Now, a new technique being developed by researchers at the Massachusetts Institute of Technology (MIT; Cambridge, MA) and elsewhere shows promise for producing highly detailed images of individual proteins, no matter how complicated their structure, without the need for crystallization.
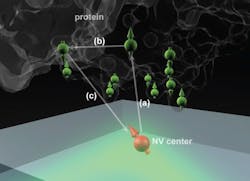
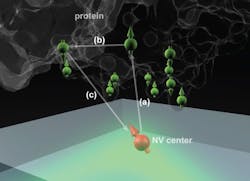
The technique makes use of microscopic defects within the crystal structure of diamond—defects that can be induced, in a controlled way, in the lab. These defects, called nitrogen vacancy (NV) centers, occur when nitrogen atoms are introduced into the crystal structure, each replacing one carbon atom in a perfectly spaced diamond lattice.
Such lattices may also include naturally occurring vacancies—imperfections where a carbon atom is missing from its normal place in the lattice. When a nitrogen atom and a vacancy come together, they form an NV center, which can be harnessed to detect the position and attributes—specifically, the spin states—of protons and electrons in atoms placed very close to them.
That's done by shining laser light at the diamond surface, which causes the NV centers to fluoresce. By detecting and analyzing the emitted light, it is possible to reconstruct details of the spin of nearby particles.
The ability to use NV centers in diamond has developed in the last few years, says MIT graduate student Ashok Ajoy, and many groups are now working to make use of them for applications in quantum computation and quantum communication. When the NV centers are very close to a diamond's surface—within a few nanometers—they can also be used to sense the spin states of particles within a molecule placed on the surface. The individual atoms and their positions can then, in principle, be detected and mapped out, revealing the molecular structure.
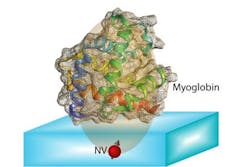
The idea is to "place a biological molecule on top of the diamond, and try to determine its structure," Ajoy explains. With proteins, "the structure and function are closely related," he says, so being able to map out that structure precisely could help in understanding both how some basic biological processes work, and how new drugs might be developed to interact with specific molecular targets.
Efforts to decode the molecular structure of proteins have mostly used x-ray crystallography, transmission electron microscopy, or nuclear magnetic resonance. But all of these methods require large sample volumes—for example, x-ray diffraction requires aggregating the molecules as crystals—so none of them can be used to study individual molecules. This greatly limits the applicability of such techniques.
What's more, while other techniques require specialized conditions such as very low temperatures or a vacuum, the new technique "perhaps can determine structure at room temperature, under ambient conditions," Ajoy says.
The next step, which the team has already begun, is to produce actual images based on this technique. "We started building this setup a year ago, and we have preliminary experiments," Ajoy says. Actual images of molecules are probably still a few years away, he says.
Full findings from the work appear in the journal Physical Review X; for more information, please visit http://dx.doi.org/10.1103/physrevx.5.011001.
-----
Follow us on Twitter, 'like' us on Facebook, connect with us on Google+, and join our group on LinkedIn
Subscribe now to BioOptics World magazine; it's free!