Super-resolution microscopy shows how cells combat drug-resistant bacteria
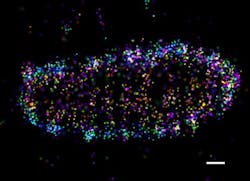
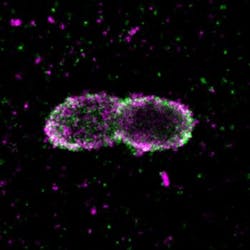
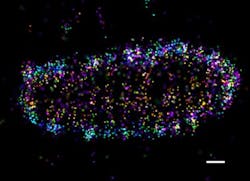
Researchers at Goethe University Frankfurt (Germany) used super-resolution microscopy to study how drug-resistant bacteria such as salmonella survive and propagate within host cells, visualizing protein patterns at the near-molecular level.
Related: Optical trapping method observes bacteria in super resolution without sample prep
Upon bacterial invasion, cells flag escaped bacteria with a small protein called ubiquitin, which is known to regulate numerous cellular processes. The attached flags contain chains of differently linked ubiquitin molecules, resulting in a secret code that has so far only partially been decoded. These ubiquitin chains relay specific signals from the surface of the bacteria into the cell.
Employing super-resolution microscopy, the research team was able to visualize different ubiquitin chains on the bacterial surface and analyze their molecular organization in detail. They discovered that one chain type—so-called linear chains—plays an essential role during a bacterial invasion. Linear ubiquitin chains trigger degradation of bacteria and kick off an inflammatory signaling cascade that results in restricting bacterial proliferation. In addition, the researchers identified the enzyme otulin as an important regulator capable of limiting this reaction—a very important notion, considering the fact that excessive inflammation is one of the major causes of tissue damage following bacterial infection.
Signaling the cells' need for pathogen defense is just one important role of ubiquitin. The small protein is also involved in development and progression of inflammatory and neurodegenerative diseases, as well as cancer. Until now, however, very little is known about how small errors in the ubiquitin system contribute to these serious human diseases, and how the system can be targeted pharmaceutically.
These new findings pave the way for many follow-up projects, which may ultimately lead to novel therapeutic approaches.
Full details of the work appear in the journal Nature Microbiology; for more information, please visit http://dx.doi.org/10.1038/nmicrobiol.2017.66.