DNA SEQUENCING/GENOMICS: Toward personalized medicine: 3G DNA sequencing
Just on the horizon, new DNA sequencing systems promise real-time processing at reduced cost with single-molecule resolution
By Stephen Turner
DNA sequencing technology has evolved through several iterations during the past decade, and the race is now on to advance the technology to a point where it makes personalized medicine feasible.
The most widely used DNA sequencing platform, the first generation technology used in the Human Genome Project, has been supplemented by so-called "next generation" technologies. Though the second generation systems have delivered dramatically higher throughput and lowered the cost per base for DNA sequencing, they have done so at the expense of some critical parameters: long read length (the number of continuous nucleotide bases read simultaneously), fast time to result, and low cost per run.
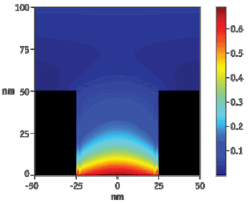
FIGURE 1. Attenuated light from the excitation beam penetrates only the lower 20-30 nm of each zero-mode waveguide (ZMW), creating a detection volume of 20 zeptoliters (10-21 liters).
For example, second generation technology has reduced read length 2 to 30 fold, increased the time to result by between 10 and 100-fold, and increased the cost per run as high as $10,000 or more.1 These performance characteristics render this generation of DNA sequencing technology incapable of resolving the true scope of medically important structural variation in the genome, addressing time-sensitive applications such as clinical diagnostics, or enabling the long tail of sequencing projects that don't rise to the current price threshold.
The result has been that while the usage patterns of first generation Sanger sequencers have evolved, the installed base and annual sales of these instruments have dropped only slightly since the advent of the second generation systems. The widely publicized goal of whole-genome sequencing as part of the delivery of personalized medicine requires not only that the limitations of existing systems be overcome, but that the costs of sequencing be reduced to a level on par with other laboratory tests. Second generation technologies have already realized much of their potential for performance improvement. A breakthrough "third generation" technology, offering the ability to sequence DNA in real-time with reduced cost, is needed to realize the promise of human genome sequencing.
How 3G is different: One example
Third generation DNA sequencing technologies are differentiated by single-molecule resolution, very long reads, fast time to results, and lower overall cost, including the flexibility to cost-effectively execute both small and large projects.
One such system is Pacific Bioscience's Single Molecule Real Time (SMRT) System. It enables a much wider range of applications when compared to second generation technologies.2 For instance, it eliminates the bottlenecks inherent in second generation technologies by using a proprietary DNA polymerase enzyme as a real-time sequencing engine.
By observing the natural process of DNA synthesis in real-time without interruption, the system capitalizes on the performance increases derived from billions of years of natural evolution. The inherent biological characteristics of the polymerase enzyme result in much longer read lengths while the speed of DNA synthesis drives fast time to results. In addition, by monitoring the enzyme in real-time, this approach provides a richer data set, including kinetic information. Together, these capabilities open new opportunities for disease research, including infectious disease studies, detection of rare variants, understanding the genomic complexity of cancer, and conducting epigenetic studies. Real-time detection is also critical to quick and efficient identification and classification of pathogens.
Three capabilities are key:
- A detector that enables single-molecule, real-time observation of individual fluorophore-labeled nucleotides with high signal-to-noise ratio.
- Phospholinked nucleotides, which enable long read lengths by producing a completely natural DNA strand through fast, accurate, and processive DNA synthesis.
- A platform that enables single-molecule, real-time detection as well as flexible run configurations and applications.
The observation window
The SMRT platform performs DNA sequencing on SMRT Cells, each of which contains arrays of thousands of zero-mode waveguides (ZMWs). A ZMW is a hole, tens of nanometers in diameter, fabricated using semiconductor manufacturing technologies using 100 nm metal film deposited on a silicon dioxide substrate.3
Each ZMW becomes a nanophotonic visualization chamber–blocking light from penetrating past just a few nanometers due to the phenomenon of waveguide cutoff well known in microwave engineering (see Fig. 1). This provides a detection volume of just ~100 zeptoliters (10-21 liters). Within each ZMW chamber, a single DNA polymerase molecule is attached to the bottom surface such that it permanently resides within the detection volume. At this volume, the activity of the polymerase, incorporating fluorescently labeled nucleotides into a growing DNA strand, can be detected amid the background due to the thousands of nearby labeled nucleotides.
Simultaneous and continuous detection occurs across all of the thousands of ZMWs on the SMRT Cell in real-time. Researchers have demonstrated this approach has the capability to produce individual, continuous sequencing reads thousands of nucleotides in length.
The role of phospholinked nucleotides
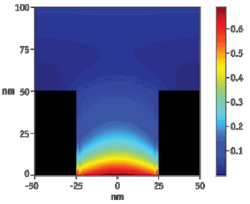
Phospholinked nucleotides, each base type labeled with a different colored fluorophore, are present in the reaction solution at high concentrations, promoting enzyme speed, accuracy, and processivity (i.e. the intrinsic read length of the polymerase).4 Due to the small size of the ZMW, even at these high concentrations, the detection volume is occupied by nucleotides only a small fraction of the time (see Fig. 2). In addition, the diffusion time, (the average duration of diffusive visitation to the detection volume) is short–just a few microseconds–due to the small distance that diffusion has to carry the nucleotides to enter and leave the detection volume. The result is a low and constant background signal.
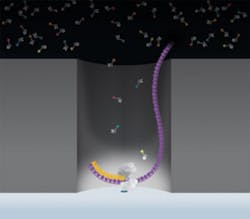
FIGURE 3. Processive synthesis with phospholinked nucleotides takes place in a series of steps: Step 1: Fluorescent phospholinked labeled nucleotides are introduced into the ZMW. Step 2: The base being incorporated is held in the detection volume for tens of milliseconds, producing a bright flash of light. Step 3: The phosphate chain is cleaved, releasing the attached dye molecule. Step 4-5: The process repeats.
As the DNA polymerase incorporates complementary nucleotides, each base is held within the detection volume for tens of milliseconds, orders of magnitude longer than the diffusion time. During this time, the polymerase-bound fluorophore emits fluorescent light, the color of which corresponds to the base identity. Then, as part of the incorporation cycle, the polymerase cleaves the bond connecting the nucleotide to the fluorophore, allowing the fluorophore to quickly diffuse out of the detection volume.
Following incorporation, the fluorescence signal returns to baseline, and the process repeats (see Fig. 3). Because of this particular way the label is attached, the DNA synthesized by the polymerase is completely natural and unmodified, leaving the polymerase without any memory of having processed an unnatural nucleotide. Inhibition of enzymatic activity, which is commonly observed with other types of labeled nucleotides, is thereby avoided.
High performance single-molecule detection
The sensitivity of the ZMW-based single-molecule detection technology is ~1,000 times greater than that of existing microscopes. This enables it to discriminate signals against background noise while reading the individual bases of DNA as close as possible to the speed in which they are synthesized in nature. A prototype instrument enables recording of labeled nucleotide incorporations by single polymerase enzymes in real-time and high multiplex.
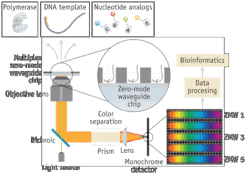
FIGURE 4. In this highly parallel optics system, a detected flash of light is separated into a spatial array–from which the identity of the incorporated base is determined.
Single-molecule fluorescence detection instrumentation dates back to the mid-1980s,5 but in a sequencing application, there are unprecedented demands on performance. First, DNA sequencing naturally implies four distinct color channels that must be simultaneously monitored. Second, the signal-to-noise ratio must be high so as to avoid missing incorporation events. And, thanks to real-time sequencing, the array of ZMWs must be monitored constantly so that incorporation events will not be missed.
A parallel optical system meets these challenges by providing more than 3,000 continuously monitored observation volumes, single-molecule emitted fluorescence sensitivity, and simultaneous spectroscopic resolution.6
Two monochromatic laser beams are divided each by wavelength-specific holographic phase masks into several thousand sub-beams propagating in a uniform rectangular array, recombined and then focused such that each ZMW nanostructure is illuminated by a single diffraction-limited spot.
Emitted fluorescence light from the labeled nucleotides is imaged through a prism assembly onto a detector array (see Fig. 4). The prism serves to disperse the emitted fluorescence, allowing a single camera to collect both spatial (ZMW position) and spectral (labeled-nucleotide type) information. A CCD camera with monolithically integrated electron-multiplication (EMCCD) and frame-transfer capability permits continuous monitoring at frame rates of hundreds of hertz. This process repeats thousands of times over the area of the CCD array, enabling the DNA sequence to be read in real-time in each ZMW across the entire SMRT Cell.
Commercial introduction
Later this year a commercial version of the detection platform with multiplex capability increased will enable monitoring of 80,000 ZMWs simultaneously and successive multiple sets of 80,000 ZMWs on the same SMRT Cell.
At initial commercial release, SMRT sequencing will be ~104 times faster than the most popular second-generation technology on a nucleotide incorporation rate basis. A single experiment can be run for as little as $99 for the SMRT Cell and sequencing reagents. The ability to run a single SMRT Cell in as little as 15 minutes, or a batch of up to 96 Cells in a single job for up to 12 hours without operator intervention, provides enormous flexibility in experimental design and implementation. This flexibility in project investment is unique compared to the long, multiday run times and resultant, single, massive data output of today's systems.
Research and collaboration promise additional applications for this platform such as simpler and more direct solutions for RNA sequencing and methylation sequencing. In addition to personalized medicine, the technology is expected to offer exciting advances in agriculture, clean energy, and global health.
References
- M.L. Metzker, 2010, "Sequencing technologies–the next generation," Nature Reviews Genetics 11:31-46.
- J. Eid et al., 2009, "Real-time dna sequencing from single polymerase molecules," Science 323:133-138.
- M. Levene et al., 2003, "Zero-mode waveguides for single-molecule analysis at high concentrations," Science 299:682-686.
- J. Korlach et al., 2008, "Long, processive enzymatic dna synthesis using 100% dye-labeled terminal phosphate-linked nucleotides," Nucleosides, Nucleotides & Nucleic Acids 27:1072-1082.
- Moerner, W. E. 2002, "A dozen years of single-molecule spectroscopy in physics, chemistry, and biophysics," J. Phys. Chem. B 106 (5): 910–927.
- P.M. Lundquist et al., 2008, "Parallel confocal detection of single molecules in real time," Optics Letters 33:1026-1028.
STEPHEN TURNER, Ph.D. is founder and Chief Technology Officer of Pacific Biosciences, Menlo Park, CA, www.pacificbiosciences.com, [email protected].
More Brand Name Current Issue Articles
More Brand Name Archives Issue Articles