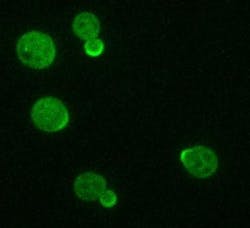
Microscopy uncovers lipid that drives cell polarity development
Researchers at the Stowers Institute for Medical Research (Kansas City, MO) have uncovered important details of the basic biochemical mechanism that establishes cell polarity in yeast. Now, using microscopy and advanced mathematics, the team has discovered that in yeast, an enzyme flips a switch by moving molecules called phospholipids from the outer to the inner layer of the cell membrane. What's more, all the molecules involved in this intricate mechanism are found not just in yeast, but also in mammalian cellsâenabling study of how cell polarity is created in liver cells, for example, and whether or not glitches in the mechanism can contribute to disease.
"Cell polarity is critical for the specialized function of the vast majority of cells," explains Rong Li, Ph.D., a professor in the Department of Molecular and Integrative Physiology at the University of Kansas Medical Center, who led the work. "Our finding could lead to a lot of clues mechanistically. We're hoping that others will pick up on our discovery and apply it to the cells they study."
Li's team uncovered an important clue to the underlying process of cell polarity. Embedded in the cell membrane is a molecule, named Cdc42, that acts as a key regulator. In a non-polarized cell, Cdc42 is randomly distributed around the membrane, like stones randomly set in a brick wall. It also floats around within the cell.
When activated, Cdc42 stimulates the formation of a skeleton of microfilaments that guide free-floating Cdc42 towards the Cdc42 already in the membrane. Some parts of the membrane start out with a bit more Cdc42 than others and the process guided by the microfilaments concentrates the molecule in those areas. Eventually, cells ends up with a high concentration of Cdc42 in one spotâthe spot where the bud will emerge.
Membrane-bound Cdc42 can diffuse away, but can also be pulled out of the membrane by another molecule named Rdi1 and then recycled back to the same spot on the membrane. The speed of the recycling determines how big the eventual spot will be, which directly affects the shape of the bud that grows out.
Faster recycling of Cdc42 results in a smaller spot on the cell; slower recycling causes a bigger spot. In a 2009 paper, Li's team showed that the recycling process can be fast or slow.
Li and her colleaguesâin seeking the answer to what controls the speed of Cdc42 dissociation and recyclingâlooked for genes that, when knocked out, affect the speed. What they found was one that slowed things way down: A type of an enzyme known as a lipid flippase, which flips lipids from one side of cell membranes to the other. Using fluorescence fluctuation spectroscopy, predoctoral researcher Arupratan Das and his colleagues were able to chart the movement of molecules in live yeast cells, and figure out the role of the flippase.
Near one end of the Cdc42 molecule is a patch with a net-positive electrical charge. The cell membrane has a negative charge. As a result, the positively charged patch of Cdc42 acts like a piece of Velcro, holding the molecule close to the membrane, allowing a lipid anchor at the end of Cdc42 to be stably inserted in the membrane. To pull out the Cdc42 for recycling, Rdi1 must grab this anchor. "Rdi1 cannot go into the membrane, so it needs the Velcro to be loosened," Li explains.
That's where the flippase enzyme comes in. The enzyme was known to flip a charge-neutral phospholipid to the inner layer of the membrane. The membrane thus becomes less negatively charged, allowing the Cdc42 anchor to slip out more easily. It's an example of how "a simple physical interaction can regulate the complex morphogenetic outcome observed during cell polarization," explains Das.
Then, Das created small bits of manmade, reconstituted membrane to test the mechanism. Using total internal reflection fluorescence (TIRF) microscopy, the team was able to show that the charge of the membrane does indeed determine how fast Cdc42 can be pulled out by Rdi1.
For more information on the team's work, which has been published in Nature Cell Biology, please visit http://www.nature.com/ncb/journal/vaop/ncurrent/full/ncb2444.html.
-----
Follow us on Twitter, 'like' us on Facebook, and join our group on LinkedIn
Follow OptoIQ on your iPhone; download the free app here.
Subscribe now to BioOptics World magazine; it's free!