PROTEOMICS/CELL BIOLOGY: In-vivo polarized fluorescence microscopy technique reveals protein arrangement
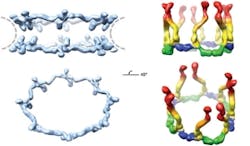
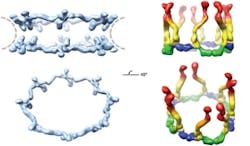
A technique based on the unique properties of polarized light has enabled scientists at Rockefeller University (New York, NY) to determine the orientation of specific proteins within cells and to make important new discoveries about the operation of the nuclear pore complex, a cluster of proteins that serves as a gateway to a cell’s nucleus.1 Although researchers have studied the nuclear pore complex for years, they have missed out on being able to visualize how the pieces fit together: Electron microscopy can reveal the broad outlines of a large protein complex, but not details. X-ray crystallography, meanwhile, can show minute detail, but only of a small piece of the complex. And anyway, both of those techniques require fixed samples, so they cannot enable visualization of cell dynamics.
After genetically attaching fluorescent markers to individual parts of the nuclear pore complex, the scientists replaced the cell’s own copy of the gene that encodes the protein with the fluorescently tagged form. Then, they used customized microscopes to measure the orientation of the waves of light the tagged proteins emitted. “Our new technique allows us to measure how components of large protein complexes are arranged in relation to one another,” says Sandy Simon, head of the Laboratory of Cellular Biophysics. “This has the potential to give us important new information about how the nuclear pore complex functions, but we believe it can also be applied to other multi-protein complexes such as those involved in DNA transcription, protein synthesis or viral replication.”
Simon told BioOptics World that his team made significant modifications to the microscopes because, “every surface that the light hit, both going into the microscope towards the sample, and then back out to the camera, distorts the polarization. Even for the simplest reflective mirrors, there were significant distortions. In the simplest case, the mirror would not reflect all orientations equally. This would obviously distort any attempts to quantify our measurements. In other cases, we would get alterations of the angle polarization.” So, he says, “each and every component had to be tested, and in most cases replaced. Finding replacements was a time-consuming task and often the more expensive mirrors were not the best.”
The scientists used the technique to study nuclear pore complexes in both budding yeast and human cells. In the case of the human cells, their new data shows that multiple copies of a key building block of the nuclear pore complex, the Y-shaped subcomplex, are arranged head-to-tail, confirming a model proposed by Blobel in 2007 that was heretofore unconfirmed. The researchers hope that the technique will eventually enable them to assemble individual crystal structures into a high-resolution map of the entire nuclear pore complex.
Because the proteins’ fluorescence can be measured while the cells are still alive, it could also give scientists new insights into how protein complexes react to varying environmental conditions, and how their configurations change over time. The approach is synergistic with other studies being conducted at Rockefeller, including analysis with X-ray crystallography, as well as electron microscopy and computer analysis—enabling a more precise picture of these complexes than has ever been possible.
1. A.L. Mattheyses et al., Biophys. J. 99 (6): 1706–1717 (2010).
More BioOptics World Current Issue Articles
More BioOptics World Archives Issue Articles