Biologists create bright-red fluorescent protein
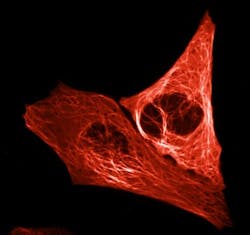
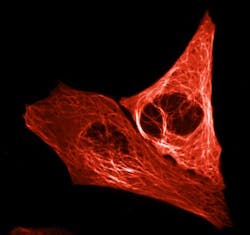
A team of biologists at the University of Amsterdam (UvA; Netherlands), working with the Institut de Biologie Structurale and the European Synchrotron Radiation Facility (ESRF; both in Grenoble, France), has created an extremely bright-red fluorescent protein for use in tracking essential cellular processes.
Related: New fluorescent protein could improve super-resolution microscopy of live cells
The bright-red fluorescent protein, dubbed mScarlet, was developed by UvA professor of molecular cytology Dorus Gadella and two of his doctoral researchers, Daphne Bindels and Lindsay Haarbosch. The group expects mScarlet to be used, for example, to gain a better understanding of how disruption of cellular processes causes uncontrolled cell division found in cancer cells.
The research team created mScarlet by comparing the genetic blueprints of a range of red fluorescent proteins from corals. They searched for sequences that consistently occurred in the various genetic codes, followed by assembling these pieces of code and then having a company synthesize a complete DNA strand. They introduced that synthetic DNA into a bacterium, which converted it into a protein.
The researchers assessed the brightness of each protein produced in this way under a microscope and then tinkered some more with the DNA code, observing how modifications affected the brightness. The entire process resulted in creating mScarlet, whose brightness will serve cellular microscopy well because it ensures the visibility of the proteins studied by scientists. Moreover, mScarlet is an ideal illuminating agent, as it does not affect the functioning of the proteins to which it is tagged.
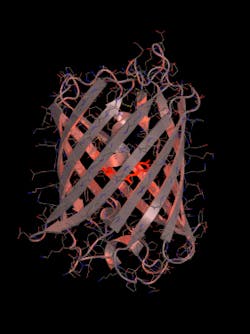
To fully understand mScarlet, the research team sent it to the Institut de Biologie Structurale. There, a team of researchers, led by structural biologist Antoine Royant, utilized the ESRF (one of the most powerful particle accelerators in the world) to reveal the molecular structure of the protein.
Full details of the work appear in the journal Nature Methods; for more information, please visit http://dx.doi.org/10.1038/nmeth.4074.