New super-resolution microscopy method is possible with single objective
Researchers at the Mechanobiology Institute (MBI) at the National University of Singapore have developed an improved super-resolution microscopy method that requires only a single objective. The method can identify single proteins anywhere within a cell and allows cellular organization to be assessed in 3D.
Related: Super-resolution microscopy visualizes the genome at the nanoscale
Recent advances in microscopy have allowed scientists to image biological samples in 3D for extended periods of time, without causing damage to the sample. Combined with super-resolution imaging, this means that the activity of single proteins can be followed within individual cells or tissues, providing new insight into protein function and, importantly, how protein dysregulation can lead to disease. Unfortunately, these imaging techniques are still prohibitively complicated and expensive for most labs.
Despite recent progress in the development of super-resolution microscopy, only a few techniques—such as total internal reflection fluorescence (TIRF) illumination and interferometric photoactivated localization microscopy (iPALM)—enable single-molecule imaging. These techniques are limited to capturing 2D images near the surface of the glass coverslip, or 3D images within the first micrometer of the coverslip. A typical human skin cell is 30 μm in height.
Selective plane illumination microscopy (SPIM) is a technique that enables 3D super-resolution imaging of thicker samples at a single-cell level. This technique selectively illuminates a single plane of the specimen by directing a focused light sheet from one side while capturing the fluorescence signal through a second objective positioned perpendicularly to the light sheet. A 3D image is reconstructed from collected images of individual cell sections. However, this approach requires a complicated, two-objective system and special sample holder, which makes it incompatible with standard microscope systems.
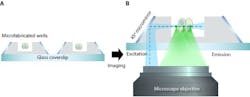
So, the researchers developed an improved SPIM method that requires only a single objective, which they call single-objective SPIM (soSPIM). The method, developed by associate professor Virgile Viasnoff, uses an array of micromirrored wells that are produced via microfabrication. Each mirror, which is inclined at precisely 45°, serves as both a means to direct the excitation beam and also to hold the sample. Together with a beam steering add-on unit, these micromirrors allow for both the excitation beam and fluorescence signal, which are viewed through the microscope, to pass through a single standard, objective lens. The soSPIM technique exhibited fast response and good sectioning capability for 3D imaging of a whole cell up to 30 μm above the coverslip. It was also able to identify single proteins deep within the cell.
With the microfabricated mirror and sample holder being produced independently of the microscope system, this technique is compatible with standard inverted microscopes and high-numerical-aperture immersion objective lenses. This will provide more researchers the ability to monitor the activity of single proteins on their existing microscope systems.
Full details of the work appear in the journal Nature Methods; for more information, please visit http://dx.doi.org/10.1038/nmeth.3402.