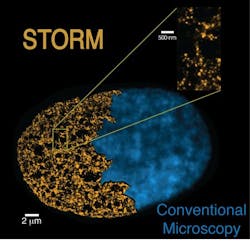
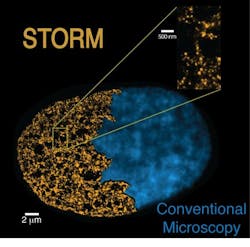
Super-resolution microscopy visualizes the genome at the nanoscale
Using a super-resolution microscopy technique, a team of scientists at the Centre for Genomic Regulation (CRG) and The Institute of Photonic Sciences (ICFO; both in Barcelona, Spain) has been able to visualize and count the smallest units that, packaged together, form the genome. In combination with innovative quantitative approaches and numerical simulations, they were also able to define the genome architecture at the nanoscale. Most importantly, they found that the nucleosomes are assembled in irregular groups across the chromatin and nucleosome-free DNA regions separate these groups.
Related: Nobel Prize honors super-resolution microscopy pioneers
Stochastic optical reconstruction microscopy (STORM), the technique that the research team used, overcomes the diffraction limit that normally restricts the spatial resolution of conventional microscopes. It also enabled the team to precisely define the chromatin fiber structure, according to Prof. Melike Lakadamyali, group leader at ICFO.
The STORM technique allowed the researchers to go deeper and, by comparing stem cells to differentiated cells (specialized cells that have already acquired their role), they observed key differences in the chromatin fiber architectures of both cells. Pia Cosma, group leader and Catalan Institution for Research and Advanced Studies (ICREA) research professor at the CRG, explains, "We found that stem cells have a different chromatin structure than somatic (specialized) cells. At the same time, this difference correlates with the level of pluripotency. The more pluripotent a cell is, the less dense is its packaging. It gives us new clues to understand the stem cells' functioning and their genomic structure, which will be helpful, for example, in studying cell reprogramming."
What scientists have found is that DNA is not regularly packaged with nucleosomes; instead, nucleosomes are assembled in groups of varying sizes—called "nucleosome clutches"—because of their similarity to egg clutches. They found that pluripotent stem cells have, on average, clutches with less density of nucleosomes. In addition, clutch size is related to the pluripotency potential of stem cells, meaning that the more pluripotent a cell is, the less nucleosomes are included in its clutches.
Even though all the cells in our body have the same genetic information, they are not expressing all the genes at the same time. So, when a cell specializes, some of the DNA regions are silenced or less accessible to the molecule that reads the genome: the RNA polymerase. Depending on the specialization of the cells, different levels of DNA packaging will occur. The research team's work establishes a new understanding of how the chromatin fibre is assembled and packaged forming a specific DNA structure in every cell.
This research definitively contributes to the understanding of a novel feature of stem cells and their DNA structure, which is important for maintaining an induced pluripotent state. A joint patent has been filed by ICFO and CRG, who are now exploring business opportunities for marketing the classification of "stemness" state of cells; i.e., their degree of pluripotency. This technique could determine with single-cell sensitivity the pluripotency potential of stem cells, thus having the capacity of becoming a standard method of quality control of stem or pluripotent cells before their use in cell therapy or research in biomedicine.
Full details of the work appear in the journal Cell; for more information, please visit http://dx.doi.org/10.1016/j.cell.2015.01.054.
-----
Follow us on Twitter, 'like' us on Facebook, connect with us on Google+, and join our group on LinkedIn