INSIDE INSTRUMENTATION: Bringing structured illumination microscopy to the masses
Maybe even the first person to look at something magnified through glass wished for more—not just more magnification, but more depth, and maybe even a three-dimensional image.
The desire to see more of the three-dimensional nature of structures continues in the biological sciences. One of the most recent approaches to making a three-dimensional representation from two-dimensional imagery comes from structured illumination microscopy (SIM). Perhaps most exciting of all, this technology now extends beyond the optical expert to the basic biologist as commercial systems come on the market.
"Structured illumination microscopy is interesting with respect to the fact that it enhances the resolution beyond conventional resolution based on diffraction, and it can provide high enough temporal resolution so that you can get dynamic images of live cells," says Stephen Ross, general manager, marketing development, Nikon. So rather than the typical light microscope's spatial resolution of 200-220 nm, SIM can capture 85-100 nm distinctions. For temporal resolution, Nikon's N-SIM grabs two frames per second. "Other super-resolution microscopes—like STORM [stochastic optical reconstruction microscopy] and PALM [photo-activated localization microscopy]—can get 10 times better resolution—down to 20-30 nanometers—but it takes many minutes to get an image," says Ross. "You can only gain so much information from such a static snapshot."
For biological sciences, in particular, nature's parameters call out for better resolution from light microscopes. As Duncan McMillan, product marketing manager at Carl Zeiss MicroImaging, states: "Many components within cells—animal and plant—are right around the diffraction limit for ordinary light-microscope resolution and smaller." Although the enhanced resolution from SIM will not uncover new subcellular structures, McMillan points out that "structured illumination lets people use fluorescence-microscopy preparations with no changes to the sample's preparation."
Exploring embryos
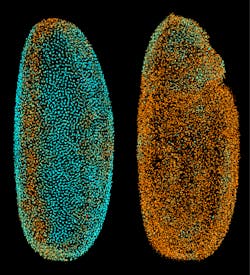
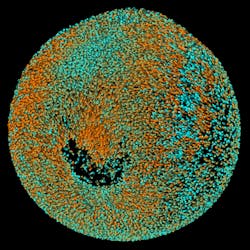
On July 4, 2010, Philipp J. Keller, a biologist who also has a background in physics and works at the Howard Hughes Medical Institute's Janelia Farm Research Campus in Ashburn, Virginia, set off a few imaging fireworks with an article he and his colleagues published online at Nature Methods. The researchers described a technique that they call digital scanned laser light-sheet fluorescence microscopy with incoherent structured illumination (DSLM-SI). To describe this technique, Keller says, "We use a sheet of light to selectively illuminate part of a specimen that we want to reconstruct in 3D." So a laser illuminates a "sheet" of the sample, excites fluorescent markers in that sheet and records the resulting fluorescence (see Fig. 1). "Then, we move the light to the next plane, and record that," Keller says.
These planes of fluorescence provide a 3D set of data. "This is very good at optical sectioning if you have a relatively transparent sample," Keller explains, "but if there's something blocking or scattering the light, then the sheet might be deformed or blur out as it propagates through the specimen." To improve on that, Keller and his colleagues added structured light patterns. "Rather than a homogenous light sheet," Keller says, "we use a periodic pattern of light and dark stripes to illuminate the specimen." Then, the same pattern can be shifted in phase, and the results can be subtracted from the original results. "You expect all scattered light to get canceled out," Keller explains, "and all that remains is the ballistic signal's contribution." The result is a high-contrast image, even from a less transparent sample.
Using DSLM-SI, Keller and his colleagues imaged a developing zebrafish over about three days (see Fig. 2). Without adding the structured light, they could only image for a day in some cases. "In some cases," Keller says, "we can now double or triple the time span of an experiment."Expanding opportunities
Luckily, biologists without a background in physics can also explore more with today's SIM. Instead of building such a microscope, researchers can buy one.
One key aspect of a commercial SIM is simplicity. In developing the Nikon N-SIM, says Ross, "We wanted a system that is built on a commercial scope. Also, it must not require an optical physicist to set it up and keep it running." In part, that required a system with fewer moving parts. For one thing, moving parts create vibrations that limit the temporal resolution of SIM. For another, fewer moving parts makes a more-robust system.
By using a commercial scope, Ross points out, the N-SIM also lets a researcher use this scope for confocal imaging, total internal reflection fluorescence and standard fluorescence. "Our system can be configured to do all of those, plus structured illumination," Ross says. To switch from one mode to another, a researcher clicks a few buttons in the user interface.
Carl Zeiss MicroImaging, too, is also releasing a SIM—called the ELYRA system. With it, a scientist can create confocal images and also use SIM and PALM technologies. This combination provides several benefits. For one thing, says McMillan, "A scientist might wonder how changing the algorithms behind SIM impacts the output image, so being able to scan the sample with confocal and then switch to SIM to compare the two is very, very helpful." He adds, "I think this will help people take the step to SIM, and you could make the same argument about the next step to PALM." Just a few mouse clicks take a researcher from one mode to the other on the ELYRA system.
Handling the data
When Keller and his colleagues started gathering data on zebrafish embryos over longer periods of time, they faced a new challenge—data. To follow an embryo over a day, Keller's device gathered 300,000 images, each being four megapixels in size and including a 16-bit dynamic range. "That makes a 2-3 terabyte dataset," Keller says. "It's too much data to go through manually." So he developed an automated analysis program. "This program converts the images to a database that you can query to follow tracks of cells, instead of going through the raw, grayscale images."
To create such a program, Keller explains that he just asked himself what a human would have to do to analyze the images. "What is a cell?" he asks. "What is a nucleus? What is an artifact? Then, I translated the answers into a computer program." He adds, "It worked reasonably well."
Still, analyzing so much data requires serious computing power. Keller started with a cluster, but it required 1,440 central processing units (CPUs) working for an entire day to reconstruct one dataset. With some adjustments, he says that the analysis can run much faster. "One of the best technical solutions is to revise the code to its fastest-running form," Keller says. "That can speed up the analysis by 10-20 times." Moreover, advanced hardware can also run the analysis faster. For example, graphical processing units (GPUs), which have been trickling from the gaming industry to scientific computing over the past few years, also speed up analysis. "GPUs are highly parallelized," Keller explains, "and they can do some of the relatively simple tasks fast, which speeds up the entire analysis quite a bit. We have already reached the point where a complete data set can be processed within a few days on a single computer workstation."
Despite the challenges of dealing with so much data, many biology labs will soon be putting SIM to work. As a result, we will soon add more structure to our understanding of how biological cells and systems work.
About the Author
Mike May
Contributing Editor, BioOptics World
Mike May writes about instrumentation design and application for BioOptics World. He earned his Ph.D. in neurobiology and behavior from Cornell University and is a member of Sigma Xi: The Scientific Research Society. He has written two books and scores of articles in the field of biomedicine.