GENOMICS/DNA SEQUENCING: DNA sequencing technologies: The next generation and beyond
There are many commonalities—and a few critical differences—among second- and third-generation sequencers. This article reveals the inner workings of each commercially available system, including specialized optical components, and explores the future of DNA sequencing.
By Elaine R. Mardis
EDITOR'S NOTE: This article was inspired by the Current Topics in Genome Analysis 2012 lecture that Dr. Mardis delivered at the National Institutes of Health.
DNA directs cell development and function, and underlies almost every aspect of human health. Since its discovery in 1953, life scientists have aimed to study it to understand gene expression in specific tissues; for instance, to learn how genome information relates to development of cancer, susceptibility to certain diseases and drug metabolism, and more.
An explosion in the ability to produce sequenced data followed completion of the human-genome sequence in 2003.1 This radical transformation has had a tremendous impact already on biological research, but its full impact has yet to be felt—because while the cost of data production has fallen dramatically, the cost of data analysis has not.
In 2004, using the only commercial sequencer available at the time—the capillary-based unit from Applied Biosystems (which later merged with Invitrogen to become Life Technologies)—sequencing the three billion base-pair human genome cost about $15 million and took weeks to years, depending upon how many sequencers were put to the task. But 6–7 years later, the cost had fallen to roughly $10,000, and at this time we are moving towards the promised $1,000 genome. Today, it takes 10–11 days to sequence six human-genome equivalents, and recent announcements of technologies have promised technology able to sequence an entire human genome overnight. And now, instead of having only one system available commercially, we have a wealth of commercially available platforms from which to choose. That's great, except that it makes for difficult decision-making.
Despite the many options, though, there are also many similarities in the offerings now available and knowing those similarities will give you a fundamental basis for understanding how the systems work (see "Shared attributes of commercially available next-gen sequencers"). So we'll review those before we compare the current crop of sequencers and look to the future, including the issue of bioinformatics.
Shared basics
Compared to the capillary approach, "next-generation" DNA sequencing systems make library creation faster, easier, and cheaper. There's no need for the time-consuming process of running through an E. coli intermediate or doing cloning; the process is straightforward, takes about a day, and begins with the random fragmentation of the starting DNA. If you're working with just polymerase chain reaction (PCR) products, then there's no need for fragmentation—you can go right to ligation with custom linkers or adapters. Then you do purification and amplification, and quantitate the library. While each instrument has its own adapters, the overall process is exactly the same and highly concordant; it takes less than a day and costs about $100–$150. The result is hundreds of millions of DNA fragments ready for sequencing.
Depending on the platform, library amplification takes place on some sort of a solid bead or glass surface—but the net impact is also the same: You're taking unique fragments and making multiple copies of each fragment as a distinct entity on that bead or glass surface in order to see the signal from the sequencing reaction. Most of these sequencers start with single molecules; they're amplified in place and then sequenced. With any type of enzymatic amplification, you'll get some biasing and some duplication or "jackpotting." This means that some library fragments will preferentially amplify, so you'll need adjustment or amelioration for that.
Massively parallel sequencing
Capillary-based approaches sequenced DNA fragments 96 or 384 at a time, then separated them by electrophoresis and used fluorescence for detection. By contrast, the new cohort of technologies incorporates direct, step-by-step detection of each nucleotide base. Commonly, these approaches are referred to as "sequencing by synthesis" if they use a polymerase, or "sequencing by ligation" if not. Perhaps a better name for these technologies is "massively parallel sequencing." This is appropriate because they sequence everything together, simultaneously, and then perform imaging to detect what happened. Thus, they don't decouple the sequencing reaction from its detection. Each step-wise base-incorporation series occurs over and over and over until the full sequence run is generated.
With a couple of exceptions, this process produces shorter reads than do capillary sequencers. There are a number of reasons for this, but the main factor is signal versus noise. This is where push comes to shove in the differential between cost to generate and cost to analyze data.
Each digital read of a massively parallel sequencer originates from one fragment in the library, which means you can literally apply counting-based methods to learn, for example, how many cells in the collective that produced DNA for a tumor genome contain each one of the mutations you've detected. That's how sensitive these systems are. You can also look at the number of counts/reads for a given messenger RNA, and at other quantitative aspects of sequencing, as we've never been able to do before. This is tremendously exciting.
Paired-end reads
One of the newest abilities of these sequencing instruments-and a function they all offer—is the use of "paired end" reads. Most of these technologies started out by priming a sequencing reaction extending off a single primer for a certain read length and delivered a single fragment read. But over time it became possible to sample from both ends of the fragments in the library. Each technology applies a different adapter to each end of the DNA fragment. You can exploit that by using one primer for one adapter, and in a second round of sequencing, another primer for the second adapter. If 300 base pairs went into the library you made, now that you have 100 from one end and 100 from the other end, you can map those to a reference genome and expect that they will align about 300 base pairs apart from one another. When that doesn't work out and they are farther apart or closer together, or maybe even mapping to separate chromosomes, this information can be interpreted as indicating a structural difference in the genome being sequenced compared to the reference sequence.
Paired-end reads have many other advantages, too, and there's a bit of a nuance involved that has become a major point of misunderstanding. There are, in fact, two kinds of paired-end reads that go by different names. In my vocabulary, "true" paired ends mean that you have a linear fragment, 300–500 base pairs, and two different primer-extension steps produce sequence data at both ends by two separate reactions. The second type of read pair that you can generate on a next-gen sequencer is a so-called "mate pair." Rather than using two separate adapters, you circularize a large DNA fragment, typically greater than 1 KB in length; 3, 8, and 20 KB libraries are typically made. By circularizing that around a common, single adapter, you actually can generate mate pairs. Where the ends come together, you go through a second step to remove the extraneous DNA, and then sequence by using the adapter or doing a single-reaction read, or by doing two separate end reads to find the sequences at either end of those fragments.
The advantage of mate pairs is that you can stretch out to much longer lengths across the DNA that you're interested in sequencing and, hopefully, better understand its long-range structure. The downside is that because DNA circularization is inherently inefficient, several micrograms of DNA are typically required for each library and multiple libraries are often needed.
But both versions more accurately place that read on the genome than is possible with a single-ended read, and this is highly advantageous when the genome is large and complex like the human genome. As long as both reads don't fall into a repetitive structure, you can anchor one with certainty, even if the other one doesn't anchor with a high degree of mapping quality. So even if a read could be placed at multiple positions in the genome, as long as its companion read places exactly at one place, it will be possible to identify just where the read came from when you align to the reference. The net result is that you can use more reads toward your analysis than with single-end reads, and this provides a huge economical advantage.
Now that we've covered the critical similarities, let's look at the differences in approaches.
Pyrosequencing: light registers incorporation
One of the first next-generation sequencing systems to come out was 454 Life Science's (Branford, CT; subsequently purchased by Roche) massively parallel version of an approach called "pyrosequencing." It uses light emission to register the incorporation of DNA nucleotides into a growing strand. It also uses the library construction approaches discussed above.
The 454 system takes a unique approach to amplification, though. Called "emulsion PCR," it generates all of the PCR in an emulsion of oil and reagents in aqueous phase. The goal is to produce a micelle containing a bead with many copies of a single DNA fragment on its surface by first isolating the bead and the originating fragment in a micelle that is filled with PCR reagents. The basic building blocks of DNA (nucleotides) and DNA polymerases will affect the amplification of this single fragment during temperature cycling, even as several million beads are being processed simultaneously in a single microtube in this emulsion.
Afterward, a series of steps breaks the emulsion to separate the oil from the water, enabling the extraction of the beads and preparing them for deposition into the sequencing plate. The 454's PicoTiter Plate is a glass structure that serves as the flow cell for the diffusion-mediated process that occurs on its upper surface (see Fig. 1). The DNA-containing beads, cleaned up from the emulsion PCR, are deposited by centrifugation into the wells-each of which accommodates one bead. Most of the wells will be filled, and that will provide a sequencing reaction when the reagents are flowed in and allowed to diffuse to enable nucleotide addition to the primer ends. The optically flat, optically clear PicoTiter Plate surface opposite the flow-cell surface abuts a high-sensitivity CCD camera that records light flashes from about a million sequencing reactions as they occur in lockstep. The pyrosequencing reaction also needs helper beads—linked with either sulfurylase or luciferase—that nestle down around the larger bead to catalyze the downstream, light-producing reactions.
The first four nucleotides for the sequencing-by-synthesis process are determined by the sequencing adapter and are always the same: A, C, T, and G. This "key sequence" calibrates the data interpretation software, telling it what a single nucleotide incorporation looks like. That's important because native nucleotides are flowing across the surface of the flow cell and in the case where, for instance, four As appear consecutively, all will be incorporated at once and the downstream output of light will be boosted to four times that of one nucleotide.
Incorporation of each nucleotide releases a pyrophosphate moiety, which participates in a series of reactions catalyzed by enzymes on the helper beads. The CCD camera detects and records the light output cycle by cycle from each PTP well. Running base incorporation cycles several hundred times to generate the read lengths from the sequencing instrument.
The newest version of the 454 instrument, the FLX Titanium+, is now approaching Sanger capillary read lengths—that is, about 650 to 700. It yields about one gigabase of data per run in about 20 hours. The error rate is about 1%; inaccuracies are typically insertion/deletion errors that occur with homopolymer runs (like the stretch of multiple As) of six or seven nucleotides (or longer)—which maxes out the detection capabilities of the camera and foils correlation to the key sequence. But sequencing isn't a one-pass process: The greater the number of sequence runs, the more opportunity you will have to get it right based on multiple samplings of the genome (also referred to as "coverage").
This 454 is a great platform for targeted validation where you're looking for single nucleotide changes because the error rate for substitutions is extraordinarily low. So if you're looking for a specific base in a PCR product, for instance, you can almost always detect it and not worry about getting a platform-specific error.
Quantity, quality, and accuracy
The Illumina (San Diego, CA) platform, another one of the early entries, was originally marketed by Solexa. Library construction occurs by a number of steps that include blunting the ends of the DNA fragments (which the 454 process also does, incidentally), phosphorylates them, and adds an A-overhang, which facilitates ligation of the adapters. Then you add clean-up and sizing steps if you want very definitive sizes from the library. In our laboratory, we've automated this to produce about 1,000 Illumina libraries a week with one technician and a fleet of small, inexpensive pipetting robots.
The surface of the "flow cell" (Illumina's name for its sequencing device) is decorated with the same type of adapters that are put onto the library fragments (see Fig. 2). This provides a point for hybridization of the single-stranded fragments. The amplification process is of a type called "bridge amplification," in which the bending DNA molecule encounters a complementary, second-end primer. The polymerase makes multiple copies in one place, generating a collection of several hundred million fragments—called a "cluster"—on the surface of the flow cell.
When the system images a cluster, it shows up as a very bright dot; imaging a portion or the entire flow cell produces what looks like a starfield. As the laser scans the incorporated fragments, they emit light that the camera detects. During the paired-end read, fragments already synthesized are washed away. Another round of amplification changes the chemistry for release and priming of the opposite end of each fragment for sequencing.
Illumina's sequencing chemistry is fundamentally different from that of the 454 in a couple of ways. For one thing, Illumina supplies all four nucleotides at each step of the nucleotide incorporation process due to the chemical aspects of the nucleotides. Each one—A, C, G, and T—has a corresponding fluor with a specific wavelength that the camera detects. In addition, at the three-prime end where normally there would be a hydroxyl group available for the next base incorporation, a chemical block is in place that disallows incorporation of another nucleotide until after the detection step, when de-blocking removes it and turns it into a hydroxyl. In a subsequent step, the fluorescent group is also cleaved off, which removes the background noise for the next step of the incorporation.
But chemistry is never 100 percent, and two things can go wrong: For some proportion of the molecules, the block will be missing—so, for example, two nucleotides may be incorporated in a single step and put the corresponding strands out of phase with the rest of the cluster. In addition, if the fluor is missing, it will be impossible to detect the molecule that's been incorporated. This may not be a problem because there are several million copies to act as backup, but if cleavage of the fluor doesn't occur, it will interfere with the signal generated in the next iteration. And because these are cumulative processes, there is plenty of room for errors—which increase noise and limit read length.
The current version of the HiSeq 2000 (with v.3 software) processes the equivalent of six human-genome sequences per run of about 10 to 11 days. Illumina recently announced the HiSeq 2500, a modification that promises dramatically less time—only about 25 hours—but also less data: about 120 gigabase pairs per run. The systems are pretty remarkable in terms of data quantity and quality and the error rate has been improving, from about 1% when we first started working with them to about 0.3% with the v3 chemistry. Version 3 also improved coverage of very high (95% and higher) G-plus-C sequences, which did not represent well in the past, and that's boosted coverage overall on the genome.
Sequencing by ligation
Finally, Life Technologies' approach-sequencing by ligation-is a different beast than the sequencing-by-polymerase approaches, although it also uses a custom adapter library approach. The instrument, called SOLiD, uses emulsion PCR on beads, and Life Technologies offers modules that facilitate the emulsion PCR process, which is otherwise manual and prone to error. The approach uses fluorescent probes about nine bases in length that have very defined content, and it primes from a common primer once the fragment-amplified beads are in the microfluidic chamber.
Rather than sequential rounds of base incorporation, this approach uses sequential rounds of ligation of primers whose defined content identifies two nucleotides in sequence at each step. The detection step involves laser excitation of the fluorescent primers that anneal according to the fragment sequences, followed by enzymatic trimming of the 9-mer to remove the fluorescent groups for the next ligation cycle. For each universal primer, multiple ligation rounds are performed. Sequencing data are obtained from sequential annealing steps with universal primers that anneal at N, N minus 1, N minus 2, etc. When the first universal primer anneals to the adapter, it's at the N-minus-zero position and the sequence is determined for bases 5, 10, 15, 20, and so on. The next time, the primer sits down at N minus 1, and the sequence is determined for bases 4, 9, etc. The beauty of this approach is that, in the design of the ligated 9-mers, the first two bases are identified with a specific known sequence, which corresponds to the fluorescent group on that particular 9-mer. So, in effect, every base is sequenced twice, because the first two nucleotides at each ligation are fixed, and these two overlap with one of the previously determined bases, effectively reading it twice. An analogy for this two-base encoding is that when you write a check, you write the digits and also write out the amount in words—which provides a cross-reference and yields an extraordinarily low substitution-error rate.
The most recent versions of the SOLiD instrument include the 5500 xl, which has an extraordinarily low error rate. It offers front-end automation, a six-lane flow chip that allows you to use only a few of the lanes if you don't have enough samples, and very high accuracy. New primer chemistries promise to increase the accuracy wildly—to 99.999%.
The third generation
The term "third-generation sequencers" refers, mostly, to a point in time—specifically the beginning of 2011 when such systems became available. Relative to the second-gen or next-gen massively parallel platforms, instruments in this class offer faster run times, lower cost per run, and a reduction in the amount of data generated. Because of these attributes, some of these systems are touting a potential to address genetic questions in a clinical setting.
This group includes Illumina's MiSeq, a scaled-down version of the HiSeq, with great similarities in terms of process and chemistry. MiSeq is a desktop instrument that works at a much lower scale.
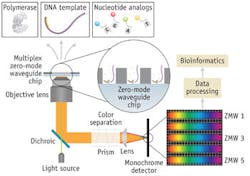
The third generation also includes the Pacific Biosciences' (Pac Bio; Menlo Park, CA) sequencer, which marries nanotechnology with molecular biology and is a true single-molecule instrument. Pac Bio uses preparatory steps very similar to those of next-gen platforms. The sample prep requires shearing, polishing of the ends, and ligation of the SMRTbell adapter, which is a clever adaptation because it is a stem-loop primer that when annealed at both fragment ends produces a circular molecule upon denaturation. The DNA polymerase and a primer that corresponds to the SMRTbell are complexed with the library molecules, which are then introduced to the surface of the SMRTcell, a nanodevice containing about 150,000 "zero-mode waveguides" (ZMWs). The DNA-polymerase complexes attach to the bottom of the ZMWs, ideally with one complex per ZMW, and fluorescent nucleotides are added. In the ZMW, the camera, optics, and laser system effectively record a real-time movie of the active site of each DNA polymerase as fluorescent nucleotides sample into the active site and are incorporated. In the process of incorporation, the fluorescent moiety diffuses away and the strand translocates, making room for the next nucleotide to float in and sample. A single run of the SMRTcell, a true real-time sequencer, involves imaging one half the chip and then the second half is imaged by shifting the laser "beamlets" that illuminate each ZMW.
Ideally, if the nucleotide is not right one, it will float out too quickly to be detected-but that is not always the case. In fact, the difficulty of single-molecule sequencing is that there's only one opportunity to get it right, and there are numerous sources of error that don't occur in amplified-molecule sequencing. Others are dark bases and multiple nucleotides getting incorporated too quickly to distinguish the individual pulses that result.
The SMRTbell is essentially a DNA lollipop that is adapted onto the ends of double-stranded fragments and gets primed with the primer—so the sequence is known. It's complementary at its ends but not in its middle, and so it forms that open, single-stranded portion of the molecule. When a denaturation impact such as sodium hydroxide is applied, the molecule opens up and becomes a circle. For very short fragments, the polymerase can sample around multiple times—both the Watson and Crick strands of that circle—going through the adapter each time. Using bioinformatics, all of the resulting sub-reads can be aligned to produce a much higher-quality consensus sequence for that short fragment.
If you want to run the sequencing for longer, you take these very large fragments (VLRs) and just sequence as long as you can during that 45-minute movie. If you're wondering, the read length is limited by data storage capacity. The movies are not stored, but rather quickly converted into a down-sampled data file that the system's software can use to do the base calling.
With the latest chemistries, read lengths can average 3,500 base pairs. But the error rate is still quite high: about 15 percent.
Strangely, most of these are insertion errors, which complicate both alignment back to a known template and the ability to assemble these reads. But we are trying to address that.
Non-optical approaches
Finally, there are two non-optical approaches worth mentioning. Life Technologies' Ion Torrent system is a variation of pyrosequencing that detects changes in pH instead of light, and works on the fact that incorporating a base releases not only a pyrophosphate, but also a hydrogen ion. Potential advantages of this approach include better sensitivity to long homopolymers because the linear dynamic range of this technology far exceeds that of a camera, and lower substitution-error rate. Over time, the Ion Torrent has been increasing in yield per run. Currently, with a two- to three-hour run time, the read length is 200 base pairs (not paired-end reads, but they're working on that) and the company is hoping, in this calendar year, to reach 400 base pairs. In September 2012, Life Technologies launched a new version of this instrument, the Ion Proton, that promises to sequence the $1,000 genome in a 24-hour period.
And, at the 2012 Advances in Genome Biology and Technology meeting, a late-breaking abstract provided by Oxford Nanopore (Oxford, England) discussed a device that could use two different processes for sequencing through nanopores. This is a substantial technological feat that many have been pursuing at the academic level for well over 15 years now, with no tangible commercial success having resulted. If these systems are real, they will revolutionize the field.
Current and future impact
The advance of DNA sequencing technology is transforming biological research. The earliest impacts have been on cancer genomics and metagenomics, but there are more on the way. Meanwhile, the clinical applications of these technologies are real, and they're pressing—we're looking now at integrating multiple data types for cancer patients. The urgency of the applications ups the ante on the extreme need for bioinformatics-based analytical approaches—not just for standard analysis, but for interpretation of the data for use by the physician—and ultimately for maximum benefit to the patient.
REFERENCE
1. E.R. Mardis, Nature, 470, 198–203 (2011).
Shared attributes of commercially available next-gen sequencers
• Random fragmentation of starting DNA, ligation with custom adapters = "a library"
• Library amplification on a solid surface (solid bead or glass)
• Direct, step-by-step detection of each nucleotide base incorporated during sequencing reaction
• Hundreds of thousands to hundreds of millions of reactions imaged per instrument run-that is, "massively parallel"
• Shorter read lengths than capillary sequencers
• A digital read type that enables direct quantitative comparisons
Elaine Mardis, Ph.D., is Professor of Genetics and Molecular Biology, as well as Co-Director and Director of Technology Development of The Genome Institute at Washington University in St. Louis School of Medicine, http://genome.wustl.edu. Contact her at http://genome.wustl.edu/people/mardis_elaine.