Raman Spectroscopy/Microscopy: Advances in SRS stimulate breakthroughs in single-cell metabolic imaging
Raman imaging is a noninvasive technique that leaves biological specimens intact and frees researchers from the need to stain them. The approach uses an excitation laser to raster-scan tissue and determine its chemical composition based on the spectroscopic properties of its component molecular bonds. Using this information, Raman imaging generates a point-by-point 3D depiction of the specimen.
While traditional Raman spectroscopy provides wonderfully specific chemical analysis, its extremely weak signal is limiting: It requires the use of high-concentration species and plenty of time, from minutes to hours.
Pioneered by Sunney Xie’s group at Harvard University, stimulated Raman scattering (SRS) improves on standard Raman techniques by amplifying the weak spontaneous Raman signal by >10,000X, which enables video-rate (30 frames/s) chemical imaging.
DO-SRS reveals developing metabolites
SRS has been developed along various lines and enabled some exciting science, including the ability to map the inner workings of living cells. Researchers at Columbia University recently reported a new SRS-based method of zooming in to track changes within individual cells, called DO-SRS. They combined SRS with the chemical tracer D2O (“heavy water”) to enable visualization of metabolic activities.1
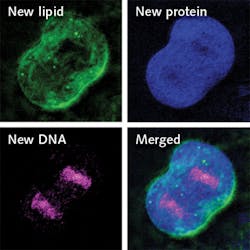
Made by swapping the hydrogen atoms in standard water with deuterium, D2O looks and tastes like regular water and is safe to drink in small doses. D2O is metabolized by cells in vivo and incorporated into newly made proteins, lipids, and DNA. With carbon, the deuterium forms chemical bonds that, as the researchers discovered, vibrate at varying frequencies when illuminated. Thus, they are able to identify macromolecules as proteins, lipids, or DNA, and also use the frequency signatures to track their growth in vivo, in the brain, skin, and other organs.
Though D2O has previously been used to label proteins and lipids for tracking metabolic changes, those methods require a mass spectrometer and extraction of cells from the body. The new method makes it possible to visualize subcellular changes in real time, in vivo. Whereas researchers have previously been able to get only a snapshot of intracellular dynamics, they can now “get a continuous picture of what’s happening inside living animal cells,” said co-lead author and postdoctoral researcher Lingyan Shi. “By tracking where and when new proteins, lipids, and DNA molecules are made, we can learn more about how animals develop and age, and what goes wrong in the case of injury and disease,” said chemistry professor Wei Min, senior author of the study.
Potential applications of the technique include quick and precise tumor removal, and detection of head injuries and developmental and metabolic disorders.
In the study, the researchers fed a mix of regular water and D2O to roundworms, mice, and zebrafish embryos, illuminated them using the SRS laser, and watched over hours and days as new deuterium-tagged proteins, lipids, and DNA grew in various tissues. The experiments revealed interesting and useful applications, including sharp delineation between healthy and cancerous tissues. In one experiment, the researchers watched cancerous cells divide in the brains and colons of mice, and emergent proteins and lipids incorporate deuterium until a bright line developed around tumors. This, Shi said, made it “much easier to remove the tumor.”
The experiments also offered numerous insights into cell development and aging. For instance, the researchers observed the formation of protein clumps in older roundworms, suggesting that DO-SRS imaging could be used to track age-related disease involving protein deposits. In the developing brains of baby mice, they observed the formation of an insulating fat layer (the myelin sheath) around each cell and determined that the new technique could help track proper brain development in children, as well as recovery in patients with multiple sclerosis, a disease that attacks the brain’s myelin and disrupts information flow.
SRP for volumetric imaging
The Columbia University work is only the latest to leverage SRS for advanced research. Other researchers are developing other SRS-based techniques, including Ji-Xin Cheng, whose team at Purdue University led work using hyperspectral SRS to learn that ovarian cancer stem cells (CSCs) contain unusually high levels of unsaturated lipids, and then demonstrated that this characteristic could serve not only as a marker for such cells, but also as a target for CSC-specific therapy.2 Now the Moustakas Chair Professor in Photonics and Optoelectronics at Boston University, Cheng is continuing development of stimulated Raman projection (SRP), a volumetric imaging technique for examining chemical species.3
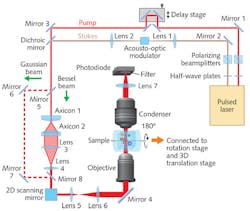
Volumetric imaging is prized for research on cell metabolism, brain function, and developmental biology because of its ability to provide quantitative molecular data on the entire volume of a complex 3D specimen. Its simplest implementation performs axial optical sectioning (which requires tightly focused laser beams), and another approach is tomography (in which a 3D model is reconstructed using images collected from multiple angles). While light-sheet fluorescence microscopy (LSFM) and optical projection tomography do image in 3D at high resolutions and speeds, they require fluorescent labels for chemical targeting, which can perturb biological functioning.Label-free SRP enables high-speed volumetric chemical imaging at a micron resolution in 3D and at high speed. In SRP, the detected signal integrates SRS intensity along the Rayleigh length of the Bessel beams (which maintain focus axially over long distances) used for input.
Compared to standard, Gaussian-beam-based SRS microscopy, Bessel-beam-based SRP microscopy can quantify the total chemical composition of a 3D volume with a two-dimensional lateral scan. Because SRP microscopy loses axial resolution, however, the team developed a corollary tomographic method. Combining SRP and sample rotation enables reconstruction of the 3D distribution of chemical compositions with optical spatial resolution at higher speeds than can Gaussian-beam-based SRS sectioning (and interestingly, greater imaging speeds in larger volumes).
Experiments on single cells confirmed that while spatial resolution was slightly lower in the SRP tomographic images than in the SRS sectional images (caused by the lower effective NA in the SRP measurement—∼0.88 in Bessel SRP vs. ∼1.07 in Gaussian SRS)—SRP tomography is indeed applicable for in vivo imaging of biomolecules in a 3D volume.
Cheng sees the technique’s usefulness for monitoring small biomolecules within cells and understanding their metabolism in situ. Based on review criteria for intellectual merit and broad impact, the National Science Foundation (NSF) awarded his team a grant in July 2018 to further develop the technique, which he plans to commercialize for broad distribution.
SRH for intraoperative histopathology
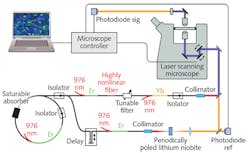
Cheng’s envisioned system may be the second species of SRS commercial instrumentation, then: Invenio Imaging, founded in 2012 and based on research led by Prof. Xie of the Department of Chemistry and Chemical Biology at Harvard University, delivered the first fully integrated SRS research microscope in 2015 to University of Michigan (UMich) researchers in the lab of Daniel Orringer, thanks to funding from the NSF Small Business Innovation Research (SBIR) Program, the National Institutes of Health (NIH) Bioengineering Research Partnership Program, and UMich’s own Translational Research and Commercialization (MTRAC) initiative. Orringer, assistant professor of neurological surgery, and his team validated the system using samples from more than 400 patients, and with funding in 2016 from the NIH SBIR program, the Invenio team developed a follow-on instrument incorporating fiber laser technology, the first-ever clinical stimulated Raman histology (SRH) imaging system. In 2017, the Congress of Neurological Surgeons (CNS) named Orringer Innovator of the Year for his role in the development of SRH.Compared to traditional intraoperative histopathologic diagnosis methods, which are both labor- and time-intensive and thus impose critical decision-making delays during brain cancer surgery, SRH enables quick, intraoperative histology of unprocessed specimens.
SRS microscopy had already demonstrated the ability to detect brain tumor infiltration quickly in fresh, unprocessed human tissues.4 In a 2017 paper written with collaborators at New York University, Brigham and Women’s Hospital, Harvard University, and the Dana-Farber Cancer Institute—in addition to UMich and Invenio Imaging—Orringer described the first application of intraoperative SRS microscopy with a portable system on unprocessed neurological specimens from 101 patients.5 The work also introduced the group’s SRH image-processing method that uses SRS images as the basis for producing virtual slides of hematoxylin-and-eosin-stained tissue, which were able to reveal essential diagnostic features. The group proved both accuracy (>92%) and specificity—concordance of SRH and conventional histology for predicting diagnosis (Cohen’s kappa, κ > 0.89). They also built and validated a machine-learning process able to predict (with 90% accuracy) brain-tumor subtype.
Gaining power
Clearly, Cheng is right to say, “SRS is becoming a very powerful tool for single-cell metabolic imaging.” As optical technologies enable systems development and as researchers push the applications, SRS will no doubt continue to increase in power and capability, further enabling the direct visualization of metabolic dynamics—in vivo, with high spatial and temporal resolution. And because this type of imaging is so essential to understanding biological processes, it promises further advances in bioscience.
REFERENCES
1. L. Shi et al., Nat. Commun., 6, 9, 1, 2995 (2018).
2. J. Li et al. Cell Stem Cell, 20, 3, 303–314 (2017).
3. X. Cheng et al. Nat. Commun., 8, 15117 (2017); doi:10.1038/ncomms15117.
4. M. Ji et al., Sci. Transl. Med., 4, 5, 201, 201ra119 (2013).
5. D. A. Orringer et al., Nat. Biomed. Eng., 1, 27 (2017).
About the Author

Barbara Gefvert
Editor-in-Chief, BioOptics World (2008-2020)
Barbara G. Gefvert has been a science and technology editor and writer since 1987, and served as editor in chief on multiple publications, including Sensors magazine for nearly a decade.