Fiberoptic arrays 'sniff out' bio threats
Researchers at Tufts University (Medford, MA) are using CCD cameras, optical-fiber bundles, and image-processing software to mimic the detection methods of the human nose. The olfactory capability of incorporating selectivity for many chemicals (broad spectral response) with high sensitivity is likely to become a useful tool in sniffing out potential bio-warfare threats, according to a presentation by Jason Epstein at the SPIE Optics and Photonics in Homeland Security conference (Dec. 11–12, 2002; Alexandria, VA).
"If you look at the nose per se, you don't have a single sensor for coffee or a sensor for apple pie," said Epstein, a member of the David Walt research group at Tufts. "You have thousands of sensors disbursed randomly in your nasal cavity and as you take a breath each sensor fires off a response. The first time you smell coffee, someone tells you it's coffee and your mind is trained to know the smell of coffee." This cross-reactive sensing method—in which complex, time-dependent signals from an array of sensors provide a signature of each analyte—yields an important potential benefit for needle-in-a-haystack detection problems in which minute quantities of target materials, such as nerve gas or mustard gas, must be quickly identified within vast and varied environments, Epstein said.
Image-processing approach
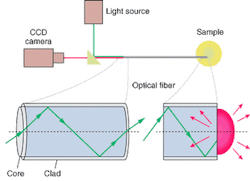
The research team has implemented this capability optically by taking advantage of the ability of each fiber in a fiberoptic bundle to transmit coherent light independently from its neighbors (see Fig. 1). "Each fiber in the bundle acts like a pixel, so a complete image is relayed from one end of the fiber to the other," Epstein said. "A typical 1-mm2 fiber bundle has about 50,000 individual 3-µm fibers fused within it, which provides 50,000 independent light pathways in which we can place an independently addressable sensor."
null
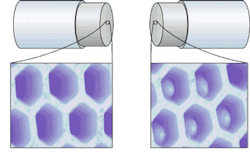
In the optical nose, polymer-immobilized dye molecules at the "sniffing" end of each fiber in a bundle provide independent fluorescent response patterns (including spectral shifts, intensity changes, spectral-shape variations, and temporal responses) in response to organic vapors, depending on the physical and chemical characteristics (such as, polarity, shape, and size) of both the vapor and the polymer.1 The actual sensors are microspheres or beads at the end of each fiber. A portion of the fiber core is etched away at the tip to leave a small pit or well. So if 3-µm wells were etched at the fiber tips, each pit could hold a 3-µm bead (see Fig. 2).
null
An excitation light source is launched down all the fibers in the bundle from the processing end. Depending on the state of the fluorescent indicator at the "sniffing" end, each fiber sends an appropriate signal back through the custom-built microscope-imaging structure onto a CCD camera for processing (see Fig. 3 and 4).2 Video images of temporal responses to various chemicals were used to successfully train a neural network for vapor recognition. In addition to fluorescent dyes for chemical sensors, the method has also been used to embed DNA-sensing microspheres, yeast cells, and E. coli cells, into fiber-tip wells of corresponding diameters, thereby creating fiber bundles specific to various sensing tasks.3, 4, 5, 6
null
Simplicity and flexibility
The latex or silica beads, all matched to the size of the etched wells and labeled with the desired sensing format, are loaded into the wells by dipping the etched fiber-bundle tips into a stock bead solution. The beads hold firmly within the wells upon drying, but can be removed through violently abrupt methods such as sonication. The relatively simple fabrication method allows simultaneous batch processing of billions of identical sensors and facilitates customization of fiber bundles for specific tasks.
"These fiber bundles provide a nice array platform, because you can actually alter existing arrays by adding new sensors as long as there are vacancies in the array," Epstein said. "You can also remove all of the beads and make a new array with new beads."
The random uptake of beads from solution into wells also boosts signal-to-noise ratios, while lowering the false-positive and false-negative rates. If the stock bead solution contains beads labeled with five different detectors, each bead type will show up in several wells at the end of a fiber bundle, and the signal-to-noise ratio improves proportional to the square root of the number of reporting sensors. False positives and false negatives are reduced by what Epstein described as a "voting scheme. If you have a dozen sensors and eleven are giving you a response indicating one thing while one is giving you a contradictory response, obviously you would go with the eleven vs. the one, which is probably misfiring."
Current projects performed by researcher Shannon Stitzel include basic olfactory research in which a three-dimensional model was created to actually illustrate the temporal olfactory sensing effect. Other research efforts are focused on detecting various potential bio-warfare agents including diseases such as anthrax and the plague, using oligonucleotide sensor arrays. The small size of the sensors enabled the researchers to detect as few as 600 copies of DNA, which is the lowest published array-based results, according to Epstein. And they are working to bring that number down even further.
"Even though our feature sizes are the smallest published for DNA arrays, we are looking to go even smaller, onto the nanoscale," he said. "And we're hoping this will give us not only faster responses and smaller sampling volumes, but a lower detection limit and higher sensitivity."
Response times are improved by the unusually small sensors, believed to be in part because the sensors exhibit radial rather than standard diffusion, and in part because the etched fiberoptic wells on the order of 3-µm examining DNA target quantities on the order of 1000 relates to micromolecular sensing concentrations, which are relatively easy to detect. Such factors further improve upon the previously mentioned signal-to-noise boost from detector redundancy.
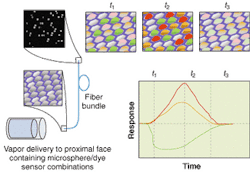
FIGURE 4. In the odor-sensing process, vacuum-controlled pulses of various vapor dilutions are delivered to the array containing the microsphere sensors (lower left). Vapor exposure induces a fluorescence intensity change at specific wavelengths, which are recorded and plotted vs. time (t1, t2, and t3). The total data produces a temporal response pattern for each sensor in the array (upper left). In this diagram, three sensor types are represented, shown as one average signal response. The colors are included to illustrate different sensor types and intensity changes.
Taking it to the next level
Despite the high DNA performance marks, Epstein described the cell-based sensing as particularly exciting. "The cells really take it to the next level because they are living," Stitzel said. "They respond to an enormous range of different analytes. And the ability to genetically engineer cells opens the doors for an enormous number of applications."
For instance, if a cell metabolizes glucose, it will form carbon dioxide and other compounds that will actually change the local pH and microenvironment of the cell, which will yield pH changes and pH responses from pH-sensitive dyes.
Further increasing the complexity, researcher Israel Biran used engineered cells with promoter and reporter genes in which the promoter responds to specific analytes and the reporter provides a fluorescent signal. Examples of reporters include green fluorescent protein and its variants, which are fluorescent biomolecules that can be found naturally in fluorescing jellyfish, for instance. Some of these genes were isolated and incorporated into a bacterial (E. coli) genome, for example. Some of the cells include promoters responsive to heavy metals like mercury, or to aromatic compounds, or anything that could be toxic, such as genotoxins that are involved in DNA damage and initiate cell SOS responses.
"You can actually go in and look at the individual response from individual cells, which is important because each cell is in a different position in its life cycle," he said. "You may have some cells that are close to death, some cells that are relatively new, some cells that are metabolically active more than others. So you can get a wide range of responses from a single assay and you can actually look at a lot of different cases." With 50,000 available wells on a fiber bundle, of course numerous multiple cell types and strains can be monitored in the same assay, indicating potential security-related applications such as high-throughput drug screening.
"The important thing about the platform is its versatility—not just each individual project, but the fact that all of the different projects are being carried out on the same platform," Epstein said. "So the key points are the optical fiber and its ability for parallel analysis."
REFERENCES
- T. A. Dickinson et al., Nature 382(22) 697 (Aug. 22, 1996).
- D. R. Walt, Science 287, 451 (Jan. 21 2000).
- S. E. Stitzel et al., Anal. Chem. 73(21) 5266 (Nov. 1, 2001).
- I. Biran, D. R. Walt, Anal. Chem. 74(13) 3046 (July 1, 2002).
- J. R. Epstein, M. Lee, D. R. Walt, Anal. Chem. 74(8) 1836 (April15, 2002).
- J. A. Ferguson, F. J. Steemers, D. R. Walt Anal. Chem. 72(22), 5618 (Nov. 15, 2000).