Image processing adds depth to cancer diagnosis
Pathologists and other biomedical researchers use confocal laser-scanning microscopy (CLSM) to examine tissue samples for cancer because it can track the genetic activity of individual cells. The method is limited, however, to viewing samples less than 100 µm thick. Hoping to boost the effectiveness of CLSM as an analytical tool, researchers at Lawrence Berkeley National Laboratory (LBNL; Berkeley, CA) have added computer analysis and control. Initial testing on breast-cancer tissue indicates a promising biomedical future for three-dimensional (3-D) microscopy.1
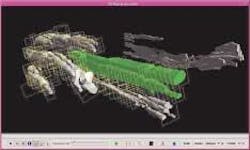
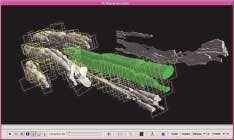
In conventional CLSM, tissue is sliced into thin segments and stained with chemical markers to highlight specific pathologies when viewed with a microscope. If pathologists want a broader picture, however, such as the extent to which cancerous cells have spread through the tissue, they must mentally stack several microscopy images together. The point of the LBNL system is to allow pathologists or other biomedical researchers to devote more of their energy to analyzing tissues for signs of disease and less to constructing three-dimensional mental images out of two-dimensional slides (see figure).
"It's something that the pathologist does in his or her brain, but we have limited capacity for reconstructing structures in three dimensions," said Carlos Ortiz de Solorzano, a staff scientist in the LBNL Life Sciences Division who helped develop the technique. "If you want to do large tissue samples you need some help. Your brain is not powerful enough to remember all of the details of the tissues, to reconstruct them in 3-D, and more important, to quantify them in 3-D.
"So we put together a system that takes images of entire tissue sections, identifies some structures of interest and reconstructs them in three dimensions."
Based on a 3-D reconstruction of the tissue morphology, the pathologist can then go back to the original slides for a higher level of detail in a specific tumor or gland of interest. "It goes both ways," Ortiz de Solorzano said. "You first want to see the organization of the structure in 3-D. Then based on your observation in 3-D, decide where you want to focus your analysis."
The LBNL system uses a client-server computer architecture in which a C-language server controls microscope functions such as acquiring and storing images, setting the exposure time of the camera, moving the stage, operating the filter wheel, automatically focusing, and acquiring multiple field-of-view images. The client application was written in Java code, however, to make it independent of a user's computer platform.1
"You can run it on a Unix workstation as we do here, or you can run it on a personal computer running Windows or Linux or any system," Ortiz de Solorzano said "It's transparent, you don't have to do any recoding or recompiling of the code."
While Java code facilitates broad use of the system independent of user platform, it is probably not the best option in terms of performance, because the microscopy images tend to be on the order of 40 Mbyte in size while Java is designed to handle the 20-kbyte-or-so images that are commonly encountered on the World Wide Web. "We're still dealing with that and making improvements to the system," Solorzano said.
They are also working on improvements to further increase the processing speed of their system and to further automate the marking and imaging process. Existing commercial and noncommercial software programs already perform some of the functions of the LBNL system, but none integrates all of the functions, Ortiz de Solorzano said. However, he expects that once their system demonstrates its usefulness, commercial vendors will become interested and bring far more resources to the problem than are available in a research laboratory.
"If we prove that this can be done in a real-time environment it could change the way that pathologists work," he said. "They are used to looking at the architecture of the tissue in a few two-dimensional sections, but we are convinced that the third dimension with full detail can be very informative and can improve the way diagnosis is done for cancer or for almost any disease in which you need to assess the structure of the tissue or of the gland. That's what keeps us working, the idea that we can make a difference in the future in the way that cancer is diagnosed."
REFERENCE
- R. Fernandez-Gonzalez et al., Microscopic Research and Techniques 59, 522 (2002).