Sequencers benefit from solid-state detectors
Sequencers benefit from solid-state detectors
Michael D. O`Neill
New optical design with a CCD detector quadruples throughput of commercial DNA sequencers.
During the past eight years, laser-based automated deoxyribonucleic acid (DNA) sequencing instruments have revolutionized molecular biology by enabling researchers to automatically determine the sequence of nucleotide bases in an already formed strand of DNA (see Laser Focus World, May 1994, p. 135). Several firms manufacture laser-based sequencers including Pharmacia Biotech (Piscataway, NJ), LI-COR (Lincoln, NE), Molecular Dynamics (Sunnyvale, CA), and Perkin-Elmer`s Applied Biosystems Division (Foster City, CA). Each manufacturer has adopted a different approach to detection of the laser-stimulated fluorescence from labeled DNA molecules (see "Other approaches to automated DNA sequencing," p. 141), but only Perkin-Elmer systems are able to simultaneously detect, from a single lane on the electrophoretic gel, emissions from all four of the fluorescent dyes (fluorophores) used to label the four bases that make up DNA.
Very-large-scale automated sequencing has assumed growing importance due, primarily, to the Human Genome Project (see Laser Focus World, June 1994, p. 16), which has as its goal the complete sequencing of all 3 ¥ 109 base pairs that make up the human genome. Among many approaches to increasing the throughput of DNA sequencers are recent advances in the optical design of a commercial system produced by Perkin-Elmer.
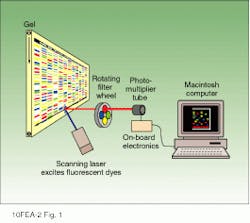
The newest Perkin-Elmer sequencer (model 377) incorporates a holographic grating spectrograph and charge-coupled-device (CCD) camera-based optical detector for simultaneous detection of four or more emission colors from the laser-excited fluorophores used to label DNA fragments. The result is an increase in overall system throughput by a factor of four. Earlier Perkin-Elmer instruments used a photomultiplier tube (PMT) for detection of the four fluorophore colors. Unlike the CCD camera, however, a PMT does not discriminate wavelengths so an instrument based on a PMT detector required four independent laser scans and a rotating wheel of wavelength-selective optical filters in order to determine the identity of each fluorophore (see Fig. 1).
Optical design improvements
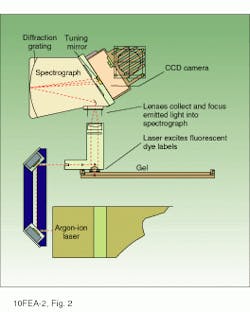
The optical configuration of the model 377 sequencer includes an argon-ion laser, focusing and collection lenses, holographic grating spectrograph, and CCD camera (see Fig. 2). The laser is a 40-mW multiline air-cooled device emitting primarily at 488 and 514.5 nm. These wavelengths excite all four fluorophores and are largely outside of the emission spectra of the dyes. The laser beam is focused to a spot size of 80 µm at the electrophoretic gel and is scanned in a horizontal line across the face of the gel at a rate of 150 mm/s.
The combined confocal laser-focusing and emission-collection optics are mounted on a moving stage; the confocal approach reduces the number of optical elements required, and consequently the number of optical adjustments is also kept to a minimum. Thus, alignment of the laser and detector is relatively easy, with the additional benefit that a confocal system is more tolerant of variables--such as the focal plane of the gel--that may be encountered while scanning the gel. Fluorescence emissions are collected and focused into the spectrograph.
In the spectrograph, the fluorescence emissions are reflected by a mirror onto a concave holographic imaging grating. This grating produces a flat-field image of the first-order spectrum at its focal plane--the CCD camera. The holographic-grating spectrograph has a dispersion of about 17 nm/mm with an "optical speed" of f/2. The slit is 1 ¥ 4 mm. The optics will focus the image to a 0.5-mm-diameter spot on the slit. The fluorescence emissions are dispersed according to wavelength into a predictably spaced pattern across the CCD camera.
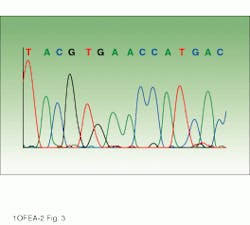
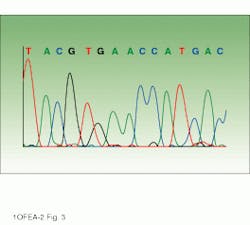
Drawing on information about which fluorophores are being used, software directs collection of the light intensities from the appropriate areas of the CCD. These software-selected areas, or virtual filters, act similarly to the fixed filters of the earlier PMT-based system. The position and width of each virtual filter can be different. Analysis software then processes the information into base sequence, fragment sizing information, or relative concentrations, depending upon the particular application. The end result is an electropherogram showing the detector response for each fluorophore (fluorescence units representing intensity) versus scan number (representing the time axis) and indicating the identity of each base (see image on p. 135).
Higher throughput is not the only benefit of the system. The CCD can be programmed for detection of specific wavelengths, which allows optimization of the signal-to-noise ratio and will make it possible for additional fluorophores to be used in future.
Applications for DNA sequencing
Incremental advances in the technology and design of DNA sequencers have steadily improved the performance of these instruments. Accelerated gene sequencing has already meant that about 40,000 of the estimated 70,000 to 100,000 human genes have been identified,1 including those associated with numerous diseases, such as cystic fibrosis, breast cancer, diabetes, obesity, polycystic kidney disease, and colon cancer.2-7 Screening tests based on DNA for these and other diseases are being developed. And DNA-based tests are used to assess the suitability of donor-recipient matches in organ/tissue transplants.
Other applications for DNA sequencing include forensic science, where it can often help confirm or deny if a piece of evidence belongs to a suspect; identification of pathogenic organisms, such as the tuberculosis organism and the plague bacillus; and identification of good breeding stock in plants and animals.
As instrument throughput increases, all these areas will be positively impacted, and completion of the Human Genome Project by the target year of 2006 will move nearer reality. In July of this year, for example, the first complete DNA sequence of a free-living organism was reported.8 This was the Haemophilus influenzae bacterium with a genome size of 1.83 ¥ 106 bases (approximately 1/1500 the size of the human genome). Theoretically, all the genes necessary for life are in this sequence, which was determined in less than a year using eight Perkin-Elmer sequencers.
Outlook
For the near term Perkin-Elmer is developing fluorescent labels and labeling reagents that will permit more than four colors to be detected in a single gel lane. The company is also collaborating with Uniphase Corp. (San Jose, CA) to develop the BioLaser, a compact low-cost source of coherent blue light specifically designed for use in DNA diagnostic instrumentation. Currently, 95% of automated DNA analysis instruments use blue-sensitive fluorescent dyes in their detection systems, and these dyes are excited by air-cooled argon lasers. Similarly, the most promising approaches currently being considered for DNA diagnostics instruments also use blue-sensitive fluorophores.
Unfortunately, the size, cost, and inefficiency of air-cooled argon-ion lasers limit development of the low-cost, compact, and convenient diagnostic instruments required for widespread adoption. Currently, the most commonly available solid-state blue/green lasers emit at 532 nm, which is not useful for the existing fluorophores used in DNA instrumentation. A small, low-cost, efficient blue light source such as the BioLaser will be an enabling advance toward realization of the next generation of DNA diagnostic instruments. n
REFERENCES
1. M. Adams et al., Nature (in press).
2. J. R. Riordan et al., "Identification of the Cystic Fibrosis Gene: Cloning and Characterization of Complementary DNA." Science 245, 1066 (1989).
3. Y. Miki et al., "A Strong Candidate for the Breast and Ovarian Cancer Susceptibility Gene BRCA1." Science 266, 66 (1994).
4. J. L. Davies et al., "A Genome-Wide Search for Human Type 1 Diabetes Susceptibility Genes." Nature 371, 130 (1994).
5. Y. Zhang et al., "Positional Cloning of the Mouse obese Gene and Its Human Homologue." Nature 372, 425 (1994).
6. M. A. Glücksmann-Kuis et al., "Polycystic Kidney Disease: The Complete Structure of the PKD1 Gene and Its Protein." Cell 81, 289 (1995).
7. R. Fishel et al., "The Human Mutator Gene Homolog MSH2 and Its Association with Hereditary Nonpolyposis Colon Cancer." Cell 75, 1027 (1993)
8. R. Fleischmann et al., "Whole-Genome Random Sequencing and Assembly of Haemophilus influenzae Rd." Science 269, 496 (1995).
DNA sequencer output can be displayed as an electropherogram or gel view. In the electropherogram (bottom), the peaks represent intensity in fluorescent units and the colors identify the four nucleotide bases, labeled A, C, G, and T. The gel view (top) displays the sequence of bases, with each base represented by one of four colors (red, blue, green, and yellow).
FIGURE 1. In the earlier Perkin-Elmer ABI Prism 373 DNA sequencer, as fluorophore-tagged DNA fragments were electro phoresed on a poly acryl amide gel past a scanning laser beam, fluorescence measurements were taken by a PMT-based detector and stored as a function of scan number (time) at four different wavelengths using four bandpass filters, each of which was used to detect one of the four fluorophores.
FIGURE 2. The scanning and CCD-based detection optics of the Perkin-Elmer Prism 377 DNA sequencer collect and analyze fluorescence emissions at four or more wavelengths simultaneously, thereby increasing the throughput of the instrument by a factor of four. Stored data are analyzed by the system`s software to provide either primary DNA sequence information or, alternatively, sizes (in base pairs), colors, and concentrations of fluorescent molecules in each sample relative to an internal fluorescent standard.
Other approaches to automated DNA sequencing
Manufacturers of DNA sequencers differ in their approaches to DNA sequencing. Three systems are described here.
nPharmacia Biotech uses a fluorescent, single-dye detection technique. The instrument incorporates a stationary 2.5-mW HeNe laser source emitting at 633 nm and 40 fixed photodetectors to analyze 10 samples simultaneously.
Laser light enters the electro phoretic gel at right angles to the direction of band migration, exciting u¥to 40 lanes (10 sequencing templates or 40 fragment samples) at a time. The photodiode receptors independently measure the light from the passing bands. Pharmacia`s one-dye chemistry eliminates problems of spectral overlap, filtering, and mobility shifts that can occur in multidye systems.
nThe system manufactured by LI-COR is based on a 785-nm semiconductor laser. Fluorescent emissions from infrared fluorescent dyes that tag the DNA strands are detected by an ultralow-noise silicon avalanche photodiode. LI-COR`s near-IR fluorescence approach provides enhanced sensitivity due to the lack of background fluorescence interference; the approach also permits use of low-cost soda-lime-glass gel plates that have low fluorescence in the IR range. The laser and detector are compact enough to be mounted on a focusing stage with confocal optics that can be moved and refocused at the point of optimal detection if the gel thickness changes or if some other variable is introduced. According to LI-COR, these sequencers offer advantages of reliability, miniaturization, and economy and are well suited to small laboratories with low-throughput requirements.
nMolecular Dynamics markets a DNA sequencer jointly with Amersham Life Science Inc., (Arlington Heights, IL). Based on a HeNe laser emitting at 594.1 nm and a Peltier-cooled photodiode array, the instrument provides fluorescent single-dye detection for eight templates in 32 lanes. Raw data is fully accessible in either a chromatogram or fluorogram format.
M. O.