OPTICAL DIAGNOSTICS: Biochemists use spectroscopic ‘fingerprints’ to spot cancers
Researchers at Northeastern University (Boston, MA) have developed a method based on vibrational microspectroscopy that automatically identifies cancer in human cells without the need for sample staining. The team, headed by Max Diem, a chemistry and chemical biology professor, regards the method as a promising alternative to visual examination for situations in which human pathologists have relatively little success.
“The idea is to be able to check cells with cancer through their chemical composition,” Diem says. “We want a completely machine-based method to diagnose cancer in excised tissues.”
In the standard diagnostic method, pathologists examine stained slides containing biopsied tissue. Usually, this identifies cancers efficiently and accurately. But in two cases the method yields too many false negatives and false positives.
“If metastases are very small, such as the micrometastases that occur when cancer moves into the lymph nodes, they are very difficult to discern visually,” Diem explains. “It’s the same when you’re looking at exfoliated human cells without accompanying tissues—in Pap smears to identify cervical cancer, for example. A cell does not walk around with a sign saying: ‘I’m cancerous.’ ”
To detect the give-away signs of cancer in those situations, the Northeastern team takes humans out of the equation. “We are shooting for a method that uses a quantifiable and quantitative method to measure cells,” Diem says. “In vibrational spectroscopy, each molecule has its own fingerprint. We’re looking at the biochemical composition of the cell, and not just a morphological change. We focus on an average chemical composition and its change with disease. The major advantage is that you can detect the disease without any markers.”
The team aims to spot the specific cellular changes that indicate the onset of cancer. That proved difficult initially because a cell’s chemistry changes over its lifetime.
“We were the first to try to understand this on a biochemical basis,” Diem recalls. “We digested certain components enzymatically out of a cell to see how spectra change. We were able to assign spectral changes to different biochemical processes.”
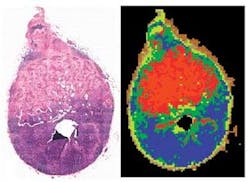
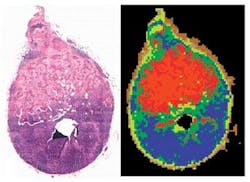
To identify those changes, the team applies infrared microscopy to biopsy slides—the same slides that pathologists use, but without any staining (see figure). “We’re mapping unstained tissues with infrared light with 6 × 6 µm pixel elements,” he says. “We reconstruct pseudo color maps like those developed for geographic images. If you compare these maps with images of stained tissue, you see just a phenomenal coincidence of the features—except that ours are created by machine.” By simply looking at a piece of tissue, they were able to see which cells are diseased and do the analysis unsupervised. The results compare well with pathologists’ findings, he adds.
What comes next? “The experiment is still too slow to be really competitive with visual cytology,” Diem admits. “But if I had $20 million lying around, I could have it on the market.” Before that can happen, of course, the method must receive approval from the U.S. Food and Drug Administration—and gain acceptance by the medical profession. But if the technology does reach the market, it could provide a significant adjunct to traditional pathology, and open new career avenues for spectroscopists.
–PG