Spectroscopy method helps decode protein used in optogenetics
Researchers at Ruhr University Bochum (RUB) and colleagues in Germany used a spectroscopy technique to help shed light upon the mode of action of the channelrhodopsin-2 protein with high spatiotemporal resolution. This protein is used in optogenetics, which is deployed to control the activity of living cells with light.
"The model we developed makes it possible to create customized optogenetic tools for individual applications," says Prof. Dr. Klaus Gerwert from the Department of Biophysics at RUB, whose team worked with Prof. Dr. Peter Hegemann's team at Humboldt University of Berlin.
Discovered by Hegemann in green algae, channelrhodopsin-2 is the central light-activated channel protein in optogenetics. If this ion channel is applied to nerve cells, the channels can be opened by light, thus activating the cell.
Gerwert explains that scientists had not been aware of what is actually happening inside channelrhodopsin-2 and what ultimately triggers its activation. But it is the understanding of processes on the atomic level that is essential for optimizing the protein specifically for its applications.
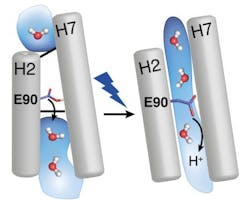
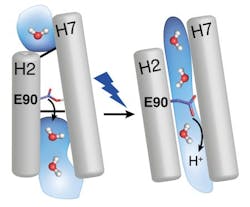
With time-resolved vibrational spectroscopy and biomolecular simulations, the research team has now closed that gap. The EHT (E90-Helix2-tilt) model describes the mode of action of channelrhodopsin-2 as follows: the light-sensitive group of the protein (the retinal) is twisted under incidence of light. This twist then continues in the protein and opens a pore quickly, which is closed by the amino acid E90 in the dark. E90 marks the narrowest place in the pore and opens it through a downward move, similar to the motion of a swing door, so that water can enter an empty vestibule above the narrowest place in the pore. The entering water then tilts the protein helix H2, which eventually triggers a protein-traversing open ion channel. When forming this model, the RUB researchers benefited from their comprehensive experience that they had gained resolving the mechanism of light-driven proton pump bacteriorhodopsin in detail.
“With this structural model, the next step, i.e. protein engineering, will become possible,” explains Gerwert. Through mutation of the amino acid E90, the protein’s properties can be controlled in a targeted manner. The conductivity or the selectivity for certain ions could be customized for specific applications, and the protein could be specifically activated with different wavelengths.
Full details of the work appear in the journal Angewandte Chemie; for more information, please visit http://dx.doi.org/10.1002/ange.201410180.
-----
Follow us on Twitter, 'like' us on Facebook, connect with us on Google+, and join our group on LinkedIn
Subscribe now to BioOptics World magazine; it's free!