DNA sequencing provides the key to the map of man
The sequencing of the human genome is the biggest watershed event in science since the lunar landing. But the potential value of this exciting news has an even bigger impact—measured in terms of life-saving drugs and a new understanding of genetic disease. All of this is made possible by a gradual evolution in biotechnology.
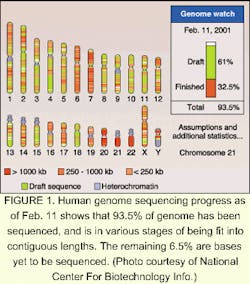
It's the biggest map yet to unfold—the sequencing of the human genome last year has opened up a whole new world of potential gain in science. On June 26, 2000, leaders of the International Human Genome Sequencing Consortium and Celera Genomics (Rockville, MD) announced the completion of a working draft of at least 93.5% of the human genome nucleotide sequence (see Fig. 1). The Human Genome Project was the culmination of 15 years of research, involving the work of more than a thousand scientists in six countries. It has opened a world of promise in pharmaceuticals and has profound implications for genetic design and the understanding of disease.
The mapping of the genome—genomics—is bringing about a revolution in our understanding of the molecular mechanisms of disease, including the complex interplay of genetic and environmental factors. Genomics is also stimulating the discovery of breakthrough healthcare products by revealing thousands of new biological targets for the development of drugs, and by giving scientists innovative ways to design new drugs, vaccines and deoxynucleic acid (DNA) diagnostics. The list of genomics-based therapeutics includes "traditional" small chemical drugs, protein drugs, and even gene therapy.
When the results were announced, the lack of genetic complexity was a surprise to many. The human genome contains 3.2 billion bases, and scientists found about 30,000 to 40,000 genes. Earlier estimates ranged from 50,000 to more than 140,000. To whit, our genetic make-up is derived from only two to three times as many genes as the fruit fly. Some 10% of our genetic sequence is clearly related to that of the fruit fly and the roundworm. Furthermore, some portions of the human sequence are shared with that of bacteria. This humbling commonality among organisms is akin to Galileo finding imperfections on the revered celestial sphere—we are not quite as unique as we think.
Follow the golden path
Often, technology evolves in small steps to bring about change with profound impact on our culture. The advent of electricity, computers, and the Internet, for example, brought about revolutions via technology that evolved gradually over many years. The technology of DNA sequencing is no different. The basic technique of gene sequencing, invented by two-time Nobel laureate Fred Sanger of the Laboratory of Molecular Biology (Cambridge, MA), has been around since the 1970s. The first automated sequencing machine was introduced in the 1980s by Leroy Hood, then at the California Institute of Technology (Pasadena, CA). In the early 1990s, the basic mapping of the genome began, involving the division of chromosomes into small, easily propagated fragments and ordering (mapping them) to the right location on the chromosome. But the real task, determining the base sequence of each of the ordered DNA fragments in a reasonably efficient way, was still elusive. By one estimate, the standard technique—gel electrophoresis—would require 30,000 work years and at least $3 billion for sequencing alone.
Improvements in DNA manipulation and chemical probes led to fluorescent dye technology, a vital step beyond radioactive probes for identifying marker bases of DNA. One breakthrough that enabled faster sequencing was capillary electrophoresis, which moves fragments of DNA simultaneously through 96 small-diameter glass capillaries for standard electrophoretic separations. These separations are much faster because the tubes dissipate heat well and allow the use of much stronger electric fields. According to Michael Phillips, section leader of instrumentation and development at Applied Biosystems (ABI; Foster City, CA), the problem to overcome was how to read the DNA at the end of all the capillaries. "The laser light is mostly lost in multiple lens effects of reflection and refraction when shone across the ends of the capillaries," explained Phillips.
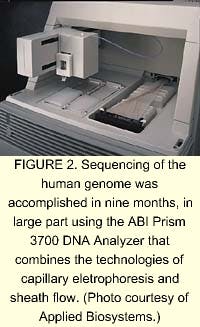
In 1999, a lightning-speed automated sequencer appeared on the horizon that combined capillary electrophoresis with a process called sheath flow. The automated DNA Analyzer developed by Applied Biosystems increased the turnaround time for fragment analysis from years to hours (see Fig. 2). Celera and the many gene-sequencing centers around the world used these DNA analyzers to sequence and analyze the human genome in about nine months (see "DNA fragment separation time drops from hours to minutes"). The technology involved in the automated gene sequencing is nothing earth shattering. The laser in the ABI analyzer was developed in the mid-1960s. The spectrograph and CCD camera have been around for decades. But combining the existing technologies with a novel technique into one good instrument may be the crowning achievement of genomics.Using 300 of ABI's automated DNA analyzers, Celera used a gene sequencing technique called whole genome shotgun (WGS) sequencing. In WGS sequencing, the DNA is first shredded into small bits of expressed genes, called expressed sequence tagsknown catalogs of expressed genes. These fluorescently labeled DNA fragments are excited at 488 and 514.5 nm by a single argon-ion laser. Sheath flow involves placing the 96 capillaries inside a cuvette, through which flows a charged polymer gel. The flow pulls the DNA fragments off the end of the capillaries, through a sieving matrix, and past the laser beam, which is directed less than 1 mm from the end of the capillaries rather than directly onto them. This procedure prevents loss of laser light from multiple lens reflection/refraction events. The charge across the flow causes the DNA to separate by fragment size. A concave spectrograph and a charge-coupled device (CCD) are used to resolve and image the four fluorescing colors of bases. The data is then translated by algorithmic processing on computers and sequenced in the proper order.
Another key achievement in sequencing involves the last step, called assembly, in which the data for the human genome is painstakingly analyzed and annotated. The field of bioinformatics, which is directed at such tasks, is a specialty of institutes such as the European Bioinformatics Institute (Hinxton, England) and the National Center for Biotechnology Information (Bethesda, MD). For assembly of the human genome, however, it was graduate student Jim Kent at the University of California, Santa Cruz, who created a program to handle the data, called GigAssembler, or "The Golden Path." GigAssembler uses an algorithm to merge overlapping fragments, and to order and orient the resulting larger fragments. To interpret the wealth of knowledge contained in the genome, biologists will be faced with the task of manipulating vast amounts of data—more than ever before. Because most biologists in the lab today are not trained in bioinformatics, mastery of the required computational methods is believed to be one of the greatest hurdles to researchers of genomic data.The not so far-flung future
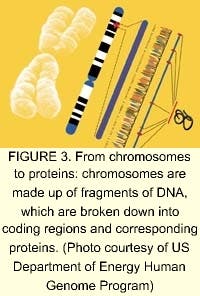
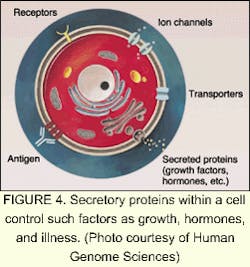
Of the tens of thousands of genes in the human genome, each one directs numerous different proteins (see Fig. 3). The study of proteins and the functions they control in the cell is an order of magnitude more complicated than the genome. According to Human Genome Sciences (Rockville, MD), most drug discovery efforts are focused on the human genes that produce signaling, or secreted, proteins that control the behavior of human organs and cells (see Fig. 4). These signaling proteins control such fundamental processes as cell growth, change, and death. Knowledge of their natural functions may allow us to help the body repair, rebuild, and restore organs and cells damaged by disease, trauma, or aging. The field of proteomics—the study of gene expression at the protein level—holds great promise for the future of medicine. Human signaling proteins, their genes, and antibodies to signaling proteins are the foundation of cutting-edge drug research (see "Facing the ethical dilemmas").The driving factor behind companies involved in proteomics and sequencing is the strong demand for DNA data. Celera plans to sell data collected from its successful sequencing of the human genome to pharmaceutical companies and research centers. In March 2000, Takeda Chemicals, a Japanese pharmaceutical manufacturer, concluded a licensing contract with Celera, allowing it to use the company's data to develop tailor-made drugs. In March 2001, Takeda signed an agreement with Human Genome Sciences for use of about 100 gene targets from its database for the same purpose.
Biotech analyst James Reddoch of Banc of America Securities (San Francisco, CA) said of the deal, "If Takeda has found 100 targets of interest, it is not unreasonable to assume that other drug companies have found similar numbers of targets." With 500 meaningful targets produced, Reddoch estimates that Human Genome Sciences could be sitting on $300 million in potential revenue from its target-sharing alliances.
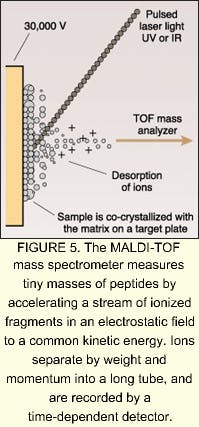
According to MRC Dunn Human Nutrition Unit (Cambridge, England), mass spectrometry is the major enabling technology for proteomics. One technology that has become prominent on the cutting edge of proteomics is matrix-assisted laser-desorption ionization time-of-flight (MALDI-TOF) mass spectrometry. In MALDI-TOF mass spectrometry, the sample is mixed in solution and allowed to co-crystallize on a target plate, forming a solid matrix. The matrix absorbs light at the excitation wavelength of a laser, which may be either a nitrogen ultraviolet system emitting at 337 nm, or an erbium:YAG laser emitting at 2.94 µm. The matrix is vaporized and accelerated toward a TOF analyzer by a voltage applied to the target plate (see Fig. 5). Flight-tube lengths are usually up to 2 m long, and flight times are about 100 ms, thousands of times longer than the nanosecond laser pulses. The sample is separated down the flight-tube by weight and mass, and detected over timed exposures. The MALDI-TOF technique is not well-suited for quantitative analyses, and resolving power and mass accuracy are limited in sequencing applications. Protein identification can be improved by use of more sophisticated mass spectrometers, such as tandem mass spectrometers. One example is the triple quadrupole (Q-TOP), which measures tryptic peptides to identify partial amino acid sequences.Many companies are hopping on the proteomics wagon. In March, for example, Shimadzu Corporation (Kyoto, Japan) launched a new strategic global business unit focused on the biotechnology and pharmaceutical sectors. The new unit, Shimadzu Biotech, plans to deliver a wide range of key products from DNA sequencers to high-performance mass spectrometry for the proteomics and genomics markets. According to Shimadzu, proteomics is rapidly becoming the focal point of biopharmaceutical research and is predicted to grow by 20% to a total of $10 billion over the next five years. According to analyst firm Business Communications (Norwalk, CT), the worldwide market for enabling biotechnologies will grow from $7.6 billion in 2000 to $17 billion by 2005.
With the genome mapped in such a surprisingly swift and sure manner, the mapping of the proteome now seems more conceivable than the lunar landing was to the Vikings.—Valerie C. Coffey
Biochips help crack the code
Microarrays and associated fluorescence-imaging equipment allow genomics researchers to carry on many DNA-sensing experiments at once.
The human race has long had the ability to manipulate genetic code. The results of such tinkering exist in the form of, for instance, the chicken, the russet potato, and the Pekinese lap dog. Late in the twentieth century, scientists learned how to bypass traditional selective-breeding techniques and insert new genes directly into the DNA of living organisms. But where to insert and what to modify in the genome remain important questions. In fact, ultimate control of the process of genetic manipulation can only come from ultimate understanding. With its much-hyped ability to lay bare the genetic code, the science of genomics is leading to better, if not ultimate, understanding of the genetic code—and a further appreciation for the complexity of life's processes.
The biochip
Central to genomics is the DNA microarray, also called the biochip. Microarrays allow scientists to test a single
segment of DNA, perhaps representing a gene, under hundreds or thousands of different conditions at once. In one type of biochip, an array of tiny samples of single-stranded probe DNA is deposited in a precise pattern, with each sample differing in genetic makeup (another type of arrangement called the macroarray works similarly, but has larger samples and spacing).
The DNA under test, also in single-stranded form, is tagged with a fluorescent marker and allowed to flow in a solution over the array. If the test DNA matches with a probe sample, it binds to that sample. Light at the proper wavelength makes the bound DNA sites fluoresce; because the DNA sequences of all the probe sites are already known, the sequence of the test DNA is revealed to the researcher by what amounts to a tiny indicator light. Such testing can determine which genes within a biological specimen are being expressed (that is, carrying out their intended function, which at a basic level is to make proteins) under a chosen experimental condition.
The use of light enters into biochip testing mainly at the fluorescence-testing stage, but also here and there at other points. Microarray-based DNA testing requires no radically new use of optoelectronics; however, components such as light sources, optics, and detectors are an important part of the test and measurement instrumentation.
Making the arrays
One of the best-known companies that produces equipment to generate DNA probe microarrays is Affymetrix (Santa Clara, CA). The company has developed a light-directed method for chemical synthesis of various DNA probes on the same chip. The technique uses photolithography in a way that is remarkably similar to that used in the construction of integrated circuits (ICs). Instead of patterning sequential thin-film layers as in the IC industry, the Affymatrix approach uses a series of photomasks and sequentially-used solutions of the four nucleotides (adenine, guanine, cytosine, and thymine, or A, G, C, and T) to build up DNA sequences one nucleotide at a time, with each probe site capable of having its own sequence.
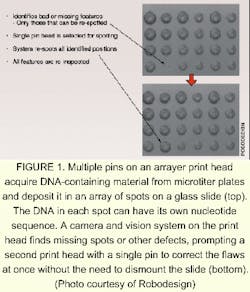
A second array-making technique relies on the deposition of an assortment of probe spots on a substrate using tiny quill pins or inkjet printing heads. Although such a system does not require an optical system to function, adding an imaging system bestows advantages. RoboDesign (Carlsbad, CA) makes an arrayer that includes a charge-coupled-device (CCD) camera and a machine-vision system. A multi-pin print head contains up to 64 quill pins, each of which picks up a dab of a different DNA-bearing solution; the print head moves to the substrate and—in the same way as does an ordinary fountain pen—deposits the dabs onto a substrate in the form of spots. The CCD camera, which is integrated onto the print head, images the spots as they are deposited. The vision system flags any spots that dry too quickly, are too small, or are otherwise substandard in quality (see Fig. 1). A single-pin rework head adjacent to the print head can quickly fix any problems, eliminating the usual procedure of dismounting the chip to perform a check, then remounting the chip.Darryl Garrison, vice president of life sciences at RoboDesign, notes that the same camera and vision system can be used to check the dimensions and quality of the pins themselves. "You just turn the print head upside down [on the instrument]," he explains. The camera and vision system can also be made part of an arrayer that uses an inkjet printing head to create DNA microarrays, he says.
Fluorescence tells all
Reading the information from a biochip requires a fluorescence-producing light source, an optical sensor to determine which DNA probe sites are fluorescing, and associated software and hardware to automate the process. Either a scanned laser spot or a broadband light field can provide the necessary illumination; the choice of light source determines the rest of the optoelectronics and mechanics in the system.
A microarray reader of the first type made by Virtek (Waterloo, Ontario, Canada) contains anywhere from two to four different types of lasers, as well as collection optics and a photomultiplier tube for detection (see photo). Information from a two-dimensional scan is used to build a fluorescence image of the microarray.The capability of scanning at more than one wavelength—and thus reading two or more dyes simultaneously—is very important, according to Reda Fayek, a technical manager at Virtek. This capability not only reduces scan times, it allows the direct comparison of, for example, DNA samples from healthy versus diseased tissue. Boosting the number of lasers to four allows two different types of test samples (each dyed with two different dyes) to be measured twice, thus creating redundancy. Alternatively, four different test samples can be measured at once. In fact, four lasers allow all four nucleotides—A, G, C, and T—to be labeled with individual dyes and identified in one experiment.
Because the scattered excitation light is many orders of magnitude higher in intensity than the fluorescence emission, creating a good fluorescence-based image requires high-quality optical filtering, says Fayek. Virtek has developed what it calls an "extreme" microarray scanning system, in which the pixel size is reduced from the usual 10 µm to 3 µm; because the image is two-dimensional, image files grow by a factor of 10, reaching a size where a compact disc can now hold only a single image file.
Fayek emphasizes that information coming back to the designer of a microarray scanner from the end-user is essential to the development of such a tool. Rather than trying to include every possible option, resulting in a scanner that is not best for any application, keeping a close watch on research trends helps the designer to emphasize what is truly important. The choice of lasers depends on the selection of usable dyes, which in turn depends on the development of fluorescent materials that tag properly to DNA. Shorter-wavelength dyes that require the use of lasers emitting in the blue are becoming available.
A microarray reader based on a broadband light source is made by Applied Precision (Issaquah, WA). White light is passed through an optical filter to select the excitation wavelength; the resulting monochromatic light is directed by a fiberoptic bundle to a ring-shaped emitting area. Fluorescent emission from the biochip passes through a second optical filter and is imaged onto a cooled high-resolution CCD camera made by Spectral Instruments (Tucson, AZ; see Laser Focus World, November 2000, p. 103). The camera collects images from small portions of the microarray, which are then stitched together in software to form one large image. Up to four excitation wavelengths in one experiment can be supported using a filter wheel. The company claims a resolution of 5 µm.
The glass bead game
Microarrays may be in the forefront when it comes to DNA analysis, but other techniques are coming into being. BioArray Solutions LLC (Piscataway, NJ) has developed a technology called light-controlled electrokinetic assembly of particles near surfaces (LEAPS), which it has adapted to DNA analysis. Rather than relying on dollops of DNA deposited directly on a substrate, LEAPS allows researchers to work with tiny glass beads coated with various DNA-probe substances. Suspended in a fluid, the beads are optically manipulated into groups on a substrate, with each group serving as its own bioreactor.
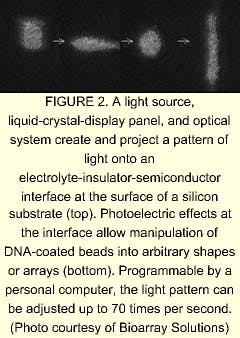
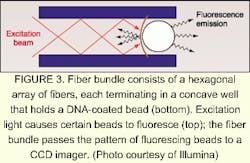
A silicon chip coated with an oxide or other insulator serving as the substrate is immersed in an electrolyte. The resulting electrolyte-insulator-semiconductor interface has electrical properties that can be modified by exposure to light. Under appropriate conditions, illuminated portions of the substrate attract or repel particles, including glass beads. The addition of a programmable illumination pattern generator creates a programmable array assembler (see Fig. 2). The apparatus permits real-time assembly of arrays of encoded beads from multiple reservoirs, with groups of beads residing in locations that reflect their reservoir of origin. Sukana Banerjee, a research manager at BioArray Solutions, explains that having many beads per array site results in the same reaction carried out many times, providing a high level of reliability through redundancy. He also notes that the LEAPS technique is not limited to glass beads—in fact, the particles to be manipulated can even be living cells.David Barker, vice president and chief scientific officer at Illumina, explains that one step of the manufacturing process is devoted to determining the type of bead at each point on the array. The decoding process consists of bringing light through the bundle at the proper excitation wavelengths in sequence and imaging the emission from the beads. The resulting information is unique to that particular fiber bundle and bead combination. In laboratory use, the beads are excited either by direct irradiation or by light through the fiber bundle. Once again, a high density of test sites per array assures redundancy. Illumina is developing a confocal imaging system to replace the CCD camera for some applications, says Barker.
As evidenced by the variety and intricacy of available DNA sensing techniques (and epitomized by the sequencing of the human genome), the field of genomics has reached a certain level of maturity. So, what is next? In addition to holding the genetic code, DNA must put such information to practical use—which it does by making proteins. The emerging field of proteomics will build on the many techniques developed for experimentation on DNA, and will likely spur the creation of new technologies, many of them based on sensing with light.—John Wallace
DNA fragment separation time drops from hours to minutes
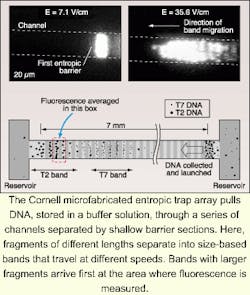
Separating DNA fragments by length is a fundamental step in DNA sequencing and therefore in the DNA fingerprinting process used to identify a person from a tissue sample or even to determine whether two people are related. Whether the resulting data is used to determine someone's predisposition to cancer or answer questions related to paternity, only within the last five years has it become possible to perform this critical sequencing step in 12 to 24 hours using gel electrophoresis. Time stands still for no one, though—which some death-row inmates waiting for a DNA-based reprieve will be relieved to hear. Hours of processing time have now become minutes with the development of a nanofabricated device that can separate double-stranded DNA fragments by length in as little as 15 to 30 minutes.
In the traditional method for separating DNA fragments, scientists place a sample at one end of a column of an organic gel and apply an electric field to the column to propel the fragments through the gel. As they slowly snake their way through the tiny pores of the material, fragments of different lengths move at different speeds. Eventually, they are collected in a series of bands as ladder-like structures that can be photographed using fluorescent or radioactive tags.
According to Harold Craighead, director of the Cornell Nanobiotechnology Center (Ithaca, NY) and colleague Jongyoon Han, their silicon-based fragment-separation prototype, which performs the same function as electrophoresis in less time, fits on a silicon chip about 15 mm long. The entropic trap array system includes channels made up of some 1400 sections, each approximately 1.5 µm deep, separated by shallow barrier sections 75 to 100 nm deep. When a DNA sample migrates through the device, fragments of different lengths travel at different speeds and arrive at the end in separate bands, with the largest fragments arriving faster than smaller ones (see figure). As with the electrophoresis process, the DNA strands are tagged with a fluorescent material, and the peaks of fluorescence at the end of the channel are measured over time to produce the data readout. Craighead and Han use an inverted microscope with a fluorescence filter set to detect the fluorescence signal from dyed DNA. Images are then were recorded and produced in video format using an integrated charge-coupled-device (ICCD) camera.In addition to being dramatically faster than gel electrophoresis, the silicon-based device can be controlled more precisely. Another advantage, according to Han, is its fabrication simplicity. Building up the silicon structure involves etching a thin channel into the substrate and then using an additional photolithography process to define thick regions in the channel. Fabrication does not require high-resolution lithography techniques because the fine dimension is controlled by a specified etch depth parameter.—Paula Noaker Powell
Facing the ethical dilemmas
The potential to predict whether individuals are predisposed to certain diseases has enormous benefits. Variations in DNA and the associated protein variants can identify, for example, known mutations in hereditary cancer genes. Patients can use such knowledge to make lifestyle changes and reduce risk factors. However, knowledge of genetic predisposition for diseases like schizophrenia, depression, or hyperactivity, could lead to privacy issues if misused by employers or health insurance providers.
In 1990, concerns such as these led the US Congress to fund the Ethical, Legal, and Social Implications (ELSI) of Human Genetics Research program. Issues to be addressed by the program include:
- to enact legislation for the protection of privacy of genetic information;
- clinical integration of genetic technologies;
- ethical research issues;
- public education;
- impact on society;
- implications to religious, philosophical and socioeconomic concerns.
Another challenging aspect of public policy is the ethics of reproductive genetic screening.
With the rapid advancement in genomics, the ELSI program will be called upon increasingly to address these issues.—Paula Noaker Powell
Sense of touch breathes life into bioresearch
Haptic devices use virtual reality to allow users to "feel" as well as see specimens.
The sensitivity of precision methods such as atomic force microscopy and optical tweezers, for viewing and probing biomolecules, is being increased, not with higher viewing resolution but by adding a sense of touch. And that sense of touch is adding a dimension of creativity for middle school and high school students as well as researchers in molecular biology.
A device that has played a major role in making this increased sensitivity possible is called a nanoManipulator and was developed by physical science researchers and computer scientists at the University of North Carolina (UNC; Chapel Hill, NC) in the early 1990s (see Laser Focus World, May 1994, p. 145). The nanoManipulator is a haptic device in that it uses virtual reality software to allow users to actually feel and probe an object or specimen that for one reason or other is not actually available to human hands, as in the case of biomolecules. The new capability has a way of turning research into a game and also of making that research more powerful.
"The main thing that you can't convey by conversation is the exhilaration of just playing with the thing," said Sean Washburn, a member of the UNC research team led by Richard Superfine in physics and Russell Taylor in computer science. "This machine permits you to do in an afternoon what prosaic step-by-step scientific-method techniques take months to do, just because you can sit down and basically ask the question, 'What's going to happen if I do this?'"
As an example, Washburn relates the story of a former University Chancellor who visited the lab and while "clumsily banging away at moving around big molecules bent one in a way that confused us." Afterwards a researcher in the field had to spend a couple of days figuring out what had happened, which led to a whole set of new experiments. Such serendipitous experimentation has become quite common, Washburn said. "Someone starts fidgeting with things and sees something unexpected."
Complementing observations
Klaus Schulten, who leads the theoretical biophysics group at the University of Illinois (Urbana, IL) and uses 3-D computer simulations and haptics for a wide range of molecular biology programs, also described a kind of productive play enabled by the haptics. He uses the UNC-developed software in his current work with atomic force microscopes and laser tweezers.
"We basically complement experimental observations," Schulten said. When experimentalists measure the mechanical response of biopolymers such as proteins, they get a limited amount of quantitative information. So the haptic measurements help them to relate the mechanical properties they measure to the actual functioning of complex biological organisms.
Schulten offered the example of titin, which he described as the longest protein in the human body in terms of its gene coding. It tends to stretch beyond its elastic limit under experimental forces in excess of 200 pico-Newtons. To help the experimentalists understand what is going on, Schulten's group first determines the complete equilibrium structure of the protein using crystallography. They can then perform computer simulations simultaneously with the experiments that provide more insight into the experimental observations.
"So we can identify the force-bearing parts of the protein," he said. "Then we can go back to the experimentalists and say, 'Why don't you mutate this part of the protein to test if we have really identified the right force-bearing component?'"
In addition to playing an increasingly wide role in investigation of biological systems, haptic devices allow computer manipulation of systems that cannot be manipulated experimentally to gain insights that could not be gained otherwise, he said.
"The most exciting thing is that we do these manipulations in real-time, meaning that while the computer is simulating the biopolymers the user sees the motion of the biopolymers in front of herself or himself on the computer screen." The user can then with a haptic device steer the device to stretch the protein or perform other operations, Schulten said. "So that's what the haptic actually does. It interacts with a computer program that makes a simulation while the system is being studied," he said. "In the old days it took several days to do a calculation, which we now do with a haptic in a few minutes."
Seeing what you're feeling
Haptics researchers at UNC are also finding plenty of work in the medical and health sciences mostly having to do with measuring the effects of friction in organic processes. One joint research project with the UNC gene therapy center involves measuring the effectiveness of delivering genomic remedies using the adenovirus. And in February, a $767,275 grant from the National Science Foundation enabled UNC researchers to make some of that work available to high school and middle school students as part of an online learning project (see Fig. 1).The basic nanoManipulator, which is now commercially available, consists of a small reverse robot arm (to provide haptic input and output) run by a desktop PC. A graphics computer or a PC with a cleverly designed graphics card provides real-time rendering of the visual images. Both the video and haptics are rendered by the same dual-processor machine. Research updates on the device at UNC include development of a microscope for use inside cells that will ultimately provide simultaneous 3-D fluorescent images and force manipulation capabilities. They are also integrating the nanoManipulator into an electron microscope, "which has the advantage that you can 'see' what you are doing as well as just 'feel' what you are doing," Washburn said.
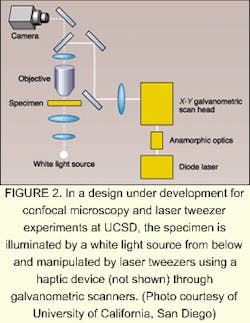
Researchers at the University of California (UCSD; San Diego, CA) are also developing an approach to 3-D reconstruction and haptics for their confocal microscopy and laser tweezers.
"Normally, real-time three-dimensional graphics involve polygon rendering, which is good for displaying precomputed surfaces at high speed given the abundance of inexpensive hardware designed specifically for rendering polygons," according to research director Jeff Squier, at UCSD. Two-dimensional pixel cross-sections provided by a digital camera in the UCSD setup and the complexity of the data sets, however, would require too much time to generate a real-time display (see Fig. 2). So, the UCSD group is rendering volume instead of polygons and is using a hardware card instead of software to perform the volume rendering.Researchers at UNC have endeavored to make the nanoManipulator available online for remote operation via Internet2, a superfast private research network. But they've run into an interesting limitation. Over distances of more than several hundred miles, the network signals are not fast enough for effective remote operation. Evidently, the immediate touch response that makes haptics so effective also makes the nanoManipulator more sensitive to time delays than devices that depend on hearing or sight. Delays of up to half a second may go unnoticed in a telephone conversation, but when delays in a haptic device increase to 1/20th of a second, users lose the sense of touch. The time lag is largely due to switching delays, which could someday be minimized, but the speed of light would ultimately provide a limit in the range of thousands of miles.—Hassaun Jones-Bey
About the Author
Valerie Coffey-Rosich
Contributing Editor
Valerie Coffey-Rosich is a freelance science and technology writer and editor and a contributing editor for Laser Focus World; she previously served as an Associate Technical Editor (2000-2003) and a Senior Technical Editor (2007-2008) for Laser Focus World.
Valerie holds a BS in physics from the University of Nevada, Reno, and an MA in astronomy from Boston University. She specializes in editing and writing about optics, photonics, astronomy, and physics in academic, reference, and business-to-business publications. In addition to Laser Focus World, her work has appeared online and in print for clients such as the American Institute of Physics, American Heritage Dictionary, BioPhotonics, Encyclopedia Britannica, EuroPhotonics, the Optical Society of America, Photonics Focus, Photonics Spectra, Sky & Telescope, and many others. She is based in Palm Springs, California.
Paula Noaker Powell
Senior Editor, Laser Focus World
Paula Noaker Powell was a senior editor for Laser Focus World.
John Wallace
Senior Technical Editor (1998-2022)
John Wallace was with Laser Focus World for nearly 25 years, retiring in late June 2022. He obtained a bachelor's degree in mechanical engineering and physics at Rutgers University and a master's in optical engineering at the University of Rochester. Before becoming an editor, John worked as an engineer at RCA, Exxon, Eastman Kodak, and GCA Corporation.
Hassaun A. Jones-Bey
Senior Editor and Freelance Writer
Hassaun A. Jones-Bey was a senior editor and then freelance writer for Laser Focus World.